
An antibody (Ab), also known as an immunoglobulin (Ig), is a large, Y-shaped protein used by the immune system to identify and neutralize foreign objects such as pathogenic bacteria and viruses. The antibody recognizes a unique molecule of the pathogen, called an antigen. Each tip of the "Y" of an antibody contains a paratope that is specific for one particular epitope on an antigen, allowing these two structures to bind together with precision. Using this binding mechanism, an antibody can tag a microbe or an infected cell for attack by other parts of the immune system, or can neutralize it directly.

Beta-lactamases, (β-lactamases) are enzymes produced by bacteria that provide multi-resistance to beta-lactam antibiotics such as penicillins, cephalosporins, cephamycins, monobactams and carbapenems (ertapenem), although carbapenems are relatively resistant to beta-lactamase. Beta-lactamase provides antibiotic resistance by breaking the antibiotics' structure. These antibiotics all have a common element in their molecular structure: a four-atom ring known as a beta-lactam (β-lactam) ring. Through hydrolysis, the enzyme lactamase breaks the β-lactam ring open, deactivating the molecule's antibacterial properties.

Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity.
Peptidoglycan or murein is a unique large macromolecule, a polysaccharide, consisting of sugars and amino acids that forms a mesh-like peptidoglycan layer outside the plasma membrane, the rigid cell wall characteristic of most bacteria. The sugar component consists of alternating residues of β-(1,4) linked N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM). Attached to the N-acetylmuramic acid is an oligopeptide chain made of three to five amino acids. The peptide chain can be cross-linked to the peptide chain of another strand forming the 3D mesh-like layer. Peptidoglycan serves a structural role in the bacterial cell wall, giving structural strength, as well as counteracting the osmotic pressure of the cytoplasm. This repetitive linking results in a dense peptidoglycan layer which is critical for maintaining cell form and withstanding high osmotic pressures, and it is regularly replaced by peptidoglycan production. Peptidoglycan hydrolysis and synthesis are two processes that must occur in order for cells to grow and multiply, a technique carried out in three stages: clipping of current material, insertion of new material, and re-crosslinking of existing material to new material.

Chymosin or rennin is a protease found in rennet. It is an aspartic endopeptidase belonging to MEROPS A1 family. It is produced by newborn ruminant animals in the lining of the abomasum to curdle the milk they ingest, allowing a longer residence in the bowels and better absorption. It is widely used in the production of cheese.
Protein engineering is the process of developing useful or valuable proteins through the design and production of unnatural polypeptides, often by altering amino acid sequences found in nature. It is a young discipline, with much research taking place into the understanding of protein folding and recognition for protein design principles. It has been used to improve the function of many enzymes for industrial catalysis. It is also a product and services market, with an estimated value of $168 billion by 2017.

Polymyxins are antibiotics. Polymyxins B and E are used in the treatment of Gram-negative bacterial infections. They work mostly by breaking up the bacterial cell membrane. They are part of a broader class of molecules called nonribosomal peptides.

Nitric oxide synthases (NOSs) are a family of enzymes catalyzing the production of nitric oxide (NO) from L-arginine. NO is an important cellular signaling molecule. It helps modulate vascular tone, insulin secretion, airway tone, and peristalsis, and is involved in angiogenesis and neural development. It may function as a retrograde neurotransmitter. Nitric oxide is mediated in mammals by the calcium-calmodulin controlled isoenzymes eNOS and nNOS. The inducible isoform, iNOS, involved in immune response, binds calmodulin at physiologically relevant concentrations, and produces NO as an immune defense mechanism, as NO is a free radical with an unpaired electron. It is the proximate cause of septic shock and may function in autoimmune disease.

Two-hybrid screening is a molecular biology technique used to discover protein–protein interactions (PPIs) and protein–DNA interactions by testing for physical interactions between two proteins or a single protein and a DNA molecule, respectively.

A leucine zipper is a common three-dimensional structural motif in proteins. They were first described by Landschulz and collaborators in 1988 when they found that an enhancer binding protein had a very characteristic 30-amino acid segment and the display of these amino acid sequences on an idealized alpha helix revealed a periodic repetition of leucine residues at every seventh position over a distance covering eight helical turns. The polypeptide segments containing these periodic arrays of leucine residues were proposed to exist in an alpha-helical conformation and the leucine side chains from one alpha helix interdigitate with those from the alpha helix of a second polypeptide, facilitating dimerization.
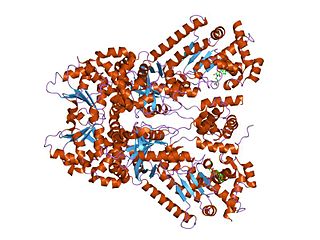
Anthrax toxin is a three-protein exotoxin secreted by virulent strains of the bacterium, Bacillus anthracis—the causative agent of anthrax. The toxin was first discovered by Harry Smith in 1954. Anthrax toxin is composed of a cell-binding protein, known as protective antigen (PA), and two enzyme components, called edema factor (EF) and lethal factor (LF). These three protein components act together to impart their physiological effects. Assembled complexes containing the toxin components are endocytosed. In the endosome, the enzymatic components of the toxin translocate into the cytoplasm of a target cell. Once in the cytosol, the enzymatic components of the toxin disrupts various immune cell functions, namely cellular signaling and cell migration. The toxin may even induce cell lysis, as is observed for macrophage cells. Anthrax toxin allows the bacteria to evade the immune system, proliferate, and ultimately kill the host animal. Research on anthrax toxin also provides insight into the generation of macromolecular assemblies, and on protein translocation, pore formation, endocytosis, and other biochemical processes.

D-amino acid oxidase is an enzyme with the function on a molecular level to oxidize D-amino acids to the corresponding α-keto acids, producing ammonia and hydrogen peroxide. This results in a number of physiological effects in various systems, most notably the brain. The enzyme is most active toward neutral D-amino acids, and not active toward acidic D-amino acids. One of its most important targets in mammals is D-Serine in the central nervous system. By targeting this and other D-amino acids in vertebrates, DAAO is important in detoxification. The role in microorganisms is slightly different, breaking down D-amino acids to generate energy.

A colicin is a type of bacteriocin produced by and toxic to some strains of Escherichia coli. Colicins are released into the environment to reduce competition from other bacterial strains. Colicins bind to outer membrane receptors, using them to translocate to the cytoplasm or cytoplasmic membrane, where they exert their cytotoxic effect, including depolarisation of the cytoplasmic membrane, DNase activity, RNase activity, or inhibition of murein synthesis.
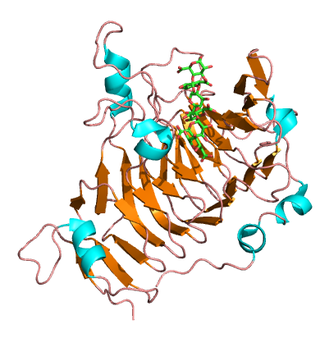
Pectinesterase (EC 3.1.1.11; systematic name pectin pectylhydrolase) is a ubiquitous cell-wall-associated enzyme that presents several isoforms that facilitate plant cell wall modification and subsequent breakdown. It catalyzes the following reaction:
Streptogramin B is a subgroup of the streptogramin antibiotics family. These natural products are cyclic hexa- or hepta depsipeptides produced by various members of the genus of bacteria Streptomyces. Many of the members of the streptogramins reported in the literature have the same structure and different names; for example, pristinamycin IA = vernamycin Bα = mikamycin B = osteogrycin B.
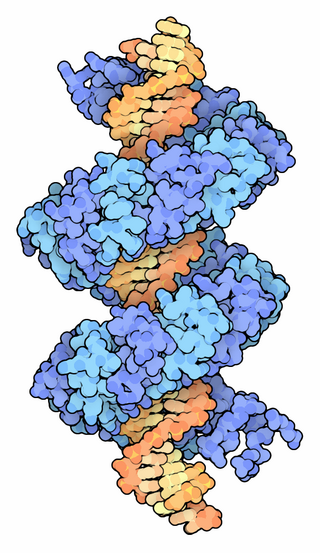
TALeffectors are proteins secreted by some β- and γ-proteobacteria. Most of these are Xanthomonads. Plant pathogenic Xanthomonas bacteria are especially known for TALEs, produced via their type III secretion system. These proteins can bind promoter sequences in the host plant and activate the expression of plant genes that aid bacterial infection. The TALE domain responsible for binding to DNA is known to have 1.5 to 33.5 short sequences that are repeated multiple times. Each of these repeats was found to be specific for a certain base pair of the DNA. These repeats also have repeat variable residues (RVD) that can detect specific DNA base pairs. They recognize plant DNA sequences through a central repeat domain consisting of a variable number of ~34 amino acid repeats. There appears to be a one-to-one correspondence between the identity of two critical amino acids in each repeat and each DNA base in the target sequence. These proteins are interesting to researchers both for their role in disease of important crop species and the relative ease of retargeting them to bind new DNA sequences. Similar proteins can be found in the pathogenic bacterium Ralstonia solanacearum and Burkholderia rhizoxinica, as well as yet unidentified marine microorganisms. The term TALE-likes is used to refer to the putative protein family encompassing the TALEs and these related proteins.

Transcription activator-like effector nucleases (TALEN) are restriction enzymes that can be engineered to cut specific sequences of DNA. They are made by fusing a TAL effector DNA-binding domain to a DNA cleavage domain. Transcription activator-like effectors (TALEs) can be engineered to bind to practically any desired DNA sequence, so when combined with a nuclease, DNA can be cut at specific locations. The restriction enzymes can be introduced into cells, for use in gene editing or for genome editing in situ, a technique known as genome editing with engineered nucleases. Alongside zinc finger nucleases and CRISPR/Cas9, TALEN is a prominent tool in the field of genome editing.
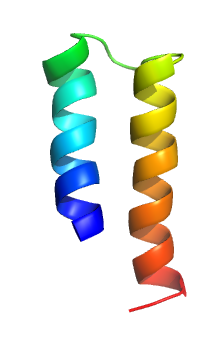
The mercury transporter superfamily is a family of transmembrane bacterial transporters of mercury ions. The common origin of all Mer superfamily members has been established. The common elements between family members are included in TMSs 1-2. A representative list of the subfamilies and proteins that belong to those subfamilies is available in the Transporter Classification Database.
Hamiltonella defensa is a species of bacteria. It is maternally or sexually transmitted and lives as an endosymbiont of whiteflies and aphids, meaning that it lives within a host, protecting its host from attack. It does this through bypassing the host's immune responses by protecting its host against parasitoid wasps. However, H. defensa is only defensive if infected by a virus. H. defensa shows a relationship with Photorhabdus species, together with Regiella insecticola. Together with other endosymbionts, it provides aphids protection against parasitoids. It is known to habitate Bemisia tabaci.

Anti-CRISPR is a group of proteins found in phages, that inhibit the normal activity of CRISPR-Cas, the immune system of certain bacteria. CRISPR consists of genomic sequences that can be found in prokaryotic organisms, that come from bacteriophages that infected the bacteria beforehand, and are used to defend the cell from further viral attacks. Anti-CRISPR results from an evolutionary process occurred in phages in order to avoid having their genomes destroyed by the prokaryotic cells that they will infect.
















