Related Research Articles

In computer science and operations research, a genetic algorithm (GA) is a metaheuristic inspired by the process of natural selection that belongs to the larger class of evolutionary algorithms (EA). Genetic algorithms are commonly used to generate high-quality solutions to optimization and search problems by relying on biologically inspired operators such as mutation, crossover and selection. Some examples of GA applications include optimizing decision trees for better performance, solving sudoku puzzles, hyperparameter optimization, causal inference, etc.
A discrete element method (DEM), also called a distinct element method, is any of a family of numerical methods for computing the motion and effect of a large number of small particles. Though DEM is very closely related to molecular dynamics, the method is generally distinguished by its inclusion of rotational degrees-of-freedom as well as stateful contact and often complicated geometries. With advances in computing power and numerical algorithms for nearest neighbor sorting, it has become possible to numerically simulate millions of particles on a single processor. Today DEM is becoming widely accepted as an effective method of addressing engineering problems in granular and discontinuous materials, especially in granular flows, powder mechanics, and rock mechanics. DEM has been extended into the Extended Discrete Element Method taking heat transfer, chemical reaction and coupling to CFD and FEM into account.

Computational biology refers to the use of data analysis, mathematical modeling and computational simulations to understand biological systems and relationships. An intersection of computer science, biology, and big data, the field also has foundations in applied mathematics, chemistry, and genetics. It differs from biological computing, a subfield of computer engineering which uses bioengineering to build computers.
Computational geometry is a branch of computer science devoted to the study of algorithms which can be stated in terms of geometry. Some purely geometrical problems arise out of the study of computational geometric algorithms, and such problems are also considered to be part of computational geometry. While modern computational geometry is a recent development, it is one of the oldest fields of computing with a history stretching back to antiquity.

Molecular dynamics (MD) is a computer simulation method for analyzing the physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a fixed period of time, giving a view of the dynamic "evolution" of the system. In the most common version, the trajectories of atoms and molecules are determined by numerically solving Newton's equations of motion for a system of interacting particles, where forces between the particles and their potential energies are often calculated using interatomic potentials or molecular mechanical force fields. The method is applied mostly in chemical physics, materials science, and biophysics.
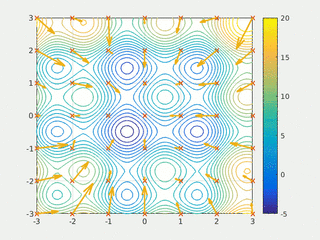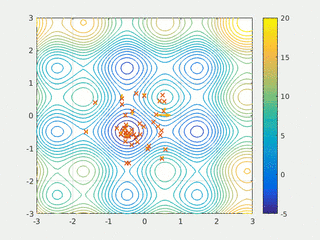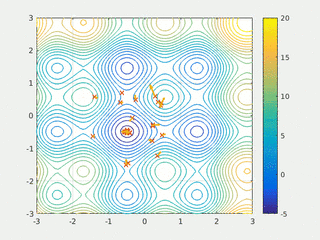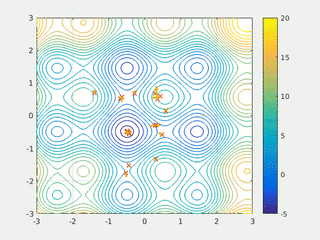
In computational science, particle swarm optimization (PSO) is a computational method that optimizes a problem by iteratively trying to improve a candidate solution with regard to a given measure of quality. It solves a problem by having a population of candidate solutions, here dubbed particles, and moving these particles around in the search-space according to simple mathematical formula over the particle's position and velocity. Each particle's movement is influenced by its local best known position, but is also guided toward the best known positions in the search-space, which are updated as better positions are found by other particles. This is expected to move the swarm toward the best solutions.

In digital image processing and computer vision, image segmentation is the process of partitioning a digital image into multiple image segments, also known as image regions or image objects. The goal of segmentation is to simplify and/or change the representation of an image into something that is more meaningful and easier to analyze. Image segmentation is typically used to locate objects and boundaries in images. More precisely, image segmentation is the process of assigning a label to every pixel in an image such that pixels with the same label share certain characteristics.
Swarm intelligence (SI) is the collective behavior of decentralized, self-organized systems, natural or artificial. The concept is employed in work on artificial intelligence. The expression was introduced by Gerardo Beni and Jing Wang in 1989, in the context of cellular robotic systems.
In parallel computing, an embarrassingly parallel workload or problem is one where little or no effort is needed to separate the problem into a number of parallel tasks. This is often the case where there is little or no dependency or need for communication between those parallel tasks, or for results between them.

A jet is a narrow cone of hadrons and other particles produced by the hadronization of a quark or gluon in a particle physics or heavy ion experiment. Particles carrying a color charge, such as quarks, cannot exist in free form because of quantum chromodynamics (QCD) confinement which only allows for colorless states. When an object containing color charge fragments, each fragment carries away some of the color charge. In order to obey confinement, these fragments create other colored objects around them to form colorless objects. The ensemble of these objects is called a jet, since the fragments all tend to travel in the same direction, forming a narrow "jet" of particles. Jets are measured in particle detectors and studied in order to determine the properties of the original quarks.
Computational genomics refers to the use of computational and statistical analysis to decipher biology from genome sequences and related data, including both DNA and RNA sequence as well as other "post-genomic" data. These, in combination with computational and statistical approaches to understanding the function of the genes and statistical association analysis, this field is also often referred to as Computational and Statistical Genetics/genomics. As such, computational genomics may be regarded as a subset of bioinformatics and computational biology, but with a focus on using whole genomes to understand the principles of how the DNA of a species controls its biology at the molecular level and beyond. With the current abundance of massive biological datasets, computational studies have become one of the most important means to biological discovery.
Reconstruction may refer to:

ParaView is an open-source multiple-platform application for interactive, scientific visualization. It has a client–server architecture to facilitate remote visualization of datasets, and generates level of detail (LOD) models to maintain interactive frame rates for large datasets. It is an application built on top of the Visualization Toolkit (VTK) libraries. ParaView is an application designed for data parallelism on shared-memory or distributed-memory multicomputers and clusters. It can also be run as a single-computer application.
Consensus clustering is a method of aggregating results from multiple clustering algorithms. Also called cluster ensembles or aggregation of clustering, it refers to the situation in which a number of different (input) clusterings have been obtained for a particular dataset and it is desired to find a single (consensus) clustering which is a better fit in some sense than the existing clusterings. Consensus clustering is thus the problem of reconciling clustering information about the same data set coming from different sources or from different runs of the same algorithm. When cast as an optimization problem, consensus clustering is known as median partition, and has been shown to be NP-complete, even when the number of input clusterings is three. Consensus clustering for unsupervised learning is analogous to ensemble learning in supervised learning.
Crystal structure prediction (CSP) is the calculation of the crystal structures of solids from first principles. Reliable methods of predicting the crystal structure of a compound, based only on its composition, has been a goal of the physical sciences since the 1950s. Computational methods employed include simulated annealing, evolutionary algorithms, distributed multipole analysis, random sampling, basin-hopping, data mining, density functional theory and molecular mechanics.
Codes for electromagnetic scattering by spheres - this article list codes for electromagnetic scattering by a homogeneous sphere, layered sphere, and cluster of spheres.
In applied mathematics, multimodal optimization deals with optimization tasks that involve finding all or most of the multiple solutions of a problem, as opposed to a single best solution. Evolutionary multimodal optimization is a branch of evolutionary computation, which is closely related to machine learning. Wong provides a short survey, wherein the chapter of Shir and the book of Preuss cover the topic in more detail.

Single particle analysis is a group of related computerized image processing techniques used to analyze images from transmission electron microscopy (TEM). These methods were developed to improve and extend the information obtainable from TEM images of particulate samples, typically proteins or other large biological entities such as viruses. Individual images of stained or unstained particles are very noisy, and so hard to interpret. Combining several digitized images of similar particles together gives an image with stronger and more easily interpretable features. An extension of this technique uses single particle methods to build up a three-dimensional reconstruction of the particle. Using cryo-electron microscopy it has become possible to generate reconstructions with sub-nanometer resolution and near-atomic resolution first in the case of highly symmetric viruses, and now in smaller, asymmetric proteins as well. Single particle analysis can also be performed by induced coupled plasma mass spectroscopy (ICP-MS).
References
- Yanting Wang, S. Teitel, and Christoph Dellago (2005), Melting of Icosahedral Gold Nanoclusters from Molecular Dynamics Simulations. Journal of Chemical Physics vol. 122, pp 214722–214738. doi : 10.1063/1.1917756