
An extremophile is an organism that is able to live in extreme environments, i.e., environments with conditions approaching or stretching the limits of what known life can adapt to, such as extreme temperature, pressure, radiation, salinity, or pH level.

Gram-negative bacteria are bacteria that unlike gram-positive bacteria do not retain the crystal violet stain used in the Gram staining method of bacterial differentiation. Their defining characteristic is their cell envelope, which consists of a thin peptidoglycan cell wall sandwiched between an inner (cytoplasmic) membrane and an outer membrane. These bacteria are found in all environments that support life on Earth.
Radioresistance is the level of ionizing radiation that organisms are able to withstand.

Burkholderia pseudomallei is a Gram-negative, bipolar, aerobic, motile rod-shaped bacterium. It is a soil-dwelling bacterium endemic in tropical and subtropical regions worldwide, particularly in Thailand and northern Australia. It was reported in 2008 that there had been an expansion of the affected regions due to significant natural disasters, and it could be found in Southern China, Hong Kong, and countries in America. B. pseudomallei, amongst other pathogens, has been found in monkeys imported into the United States from Asia for laboratory use, posing a risk that the pathogen could be introduced into the country.

Alcaligenes is a genus of Gram-negative, aerobic, rod-shaped bacteria in the order of Burkholderiales.

Ferroplasma is a genus of Archaea that belong to the family Ferroplasmaceae. Members of the Ferroplasma are typically acidophillic, pleomorphic, irregularly shaped cocci.

Cupriavidus necator is a Gram-negative soil bacterium of the class Betaproteobacteria.

Ralstonia is a genus of bacteria, previously included in the genus Pseudomonas. It is named after the American bacteriologist Ericka Ralston. Ericka Ralston was born Ericka Barrett in 1944 in Saratoga, California, and died in 2015 in Sebastopol, California. While in graduate school at the University of California at Berkeley, she identified 20 strains of Pseudomonas which formed a phenotypical homologous group, and named them Pseudomonas pickettii, after M.J. Pickett in the Department of Bacteriology at the University of California at Los Angeles, from whom she had received the strains. Later, P. pickettii was transferred to the new genus Ralstonia, along with several other species. She continued her research into bacterial pathogenesis under the name of Ericka Barrett while a professor of microbiology at the University of California at Davis from 1977 until her retirement in 1996.

Ralstonia solanacearum is an aerobic non-spore-forming, Gram-negative, plant pathogenic bacterium. R. solanacearum is soil-borne and motile with a polar flagellar tuft. It colonises the xylem, causing bacterial wilt in a very wide range of potential host plants. It is known as Granville wilt when it occurs in tobacco. Bacterial wilts of tomato, pepper, eggplant, and Irish potato caused by R. solanacearum were among the first diseases that Erwin Frink Smith proved to be caused by a bacterial pathogen. Because of its devastating lethality, R. solanacearum is now one of the more intensively studied phytopathogenic bacteria, and bacterial wilt of tomato is a model system for investigating mechanisms of pathogenesis. Ralstonia was until recently classified as Pseudomonas, with similarity in most aspects, except that it does not produce fluorescent pigment like Pseudomonas. The genomes from different strains vary from 5.5 Mb up to 6 Mb, roughly being 3.5 Mb of a chromosome and 2 Mb of a megaplasmid. While the strain GMI1000 was one of the first phytopathogenic bacteria to have its genome completed, the strain UY031 was the first R. solanacearum to have its methylome reported. Within the R. solanacearum species complex, the four major monophyletic clusters of strains are termed phylotypes, that are geographically distinct: phylotypes I-IV are found in Asia, the Americas, Africa, and Oceania, respectively.
Ralstonia pickettii is a Gram-negative, rod-shaped, soil bacterium.
The genus Massilia belongs to the family Oxalobacteriaceae, and describes a group of gram-negative, motile, rod-shaped cells. They may contain either peritrichous or polar flagella. This genus was first described in 1998, after the type species, Massilia timonae, was isolated from the blood of an immunocompromised patient. The genus was named after the old Greek and Roman name for the city of Marseille, France, where the organism was first isolated. The Massilia genus is a diverse group that resides in many different environments, has many heterotrophic means of gathering energy, and is commonly found in association with plants.
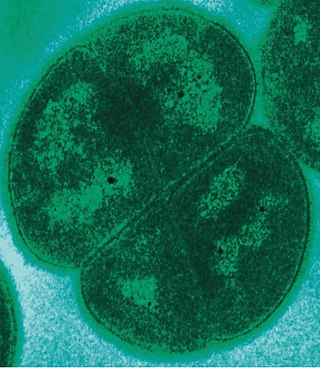
Deinococcus radiodurans is a bacterium, an extremophile and one of the most radiation-resistant organisms known. It can survive cold, dehydration, vacuum, and acid, and therefore is known as a polyextremophile. The Guinness Book Of World Records listed it as the world's toughest known bacterium.
Metallotolerants are extremophile organisms that are able to survive in environments with a high concentration of dissolved heavy metals. They can be found in environments containing arsenic, cadmium, copper, and zinc. Known metallotolerants include Ferroplasma sp. and Cupriavidus metallidurans.
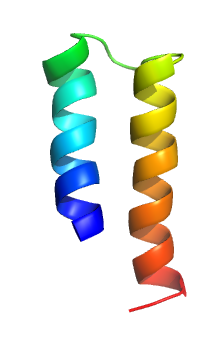
The mercury transporter superfamily is a family of transmembrane bacterial transporters of mercury ions. The common origin of all Mer superfamily members has been established. The common elements between family members are included in TMSs 1-2. A representative list of the subfamilies and proteins that belong to those subfamilies is available in the Transporter Classification Database.
Variovorax paradoxus is a gram negative, beta proteobacterium from the genus Variovorax. Strains of V. paradoxus can be categorized into two groups, hydrogen oxidizers and heterotrophic strains, both of which are aerobic. The genus name Vario-vorax and species name para-doxus (contrary-opinion) reflects both the dichotomy of V. paradoxus metabolisms, but also its ability to utilize a wide array of organic compounds.
Cupriavidus basilensis is a gram-negative soil bacterium of the genus Cupriavidus and the family Burkholderiaceae. The complete genome sequence of its type strain has been determined and is publicly available at DNA Data Bank of Japan, European Nucleotide Archive and GenBank, under the accession numbers CP062803, CP062804, CP062805, CP062806, CP062807, CP062808, CP062809 and CP062810.
Cupriavidus campinensis is a gram-negative soil bacterium of the genus Cupriavidus and the family Burkholderiaceae which was isolated in northeast Belgium. C. campinensis species were found to be highly resistant to heavy metals and antibiotics due to their genomic potentials
Cupriavidus gilardii is a Gram-negative, aerobic, motile, oxidase-positive bacterium from the genus Cupriavidus and the family Burkholderiaceae. It is motil by a single polar flagellum. It is named after G. L. Gilardi, an American microbiologist. The organism was initially identified as Ralstonia gilardii in 1999, renamed Wautersiella gilardii, and most recently moved into the genus Cupriavidus after 16S rRNA gene sequencing revealed it to be most closely related to Cupriavidus necator. Notably, species of this genus are not inhibited by copper due to the production of chelation factors, and may actually be stimulated by the presence of copper.

Resistance-nodulation-division (RND) family transporters are a category of bacterial efflux pumps, especially identified in Gram-negative bacteria and located in the cytoplasmic membrane, that actively transport substrates. The RND superfamily includes seven families: the heavy metal efflux (HME), the hydrophobe/amphiphile efflux-1, the nodulation factor exporter family (NFE), the SecDF protein-secretion accessory protein family, the hydrophobe/amphiphile efflux-2 family, the eukaryotic sterol homeostasis family, and the hydrophobe/amphiphile efflux-3 family. These RND systems are involved in maintaining homeostasis of the cell, removal of toxic compounds, and export of virulence determinants. They have a broad substrate spectrum and can lead to the diminished activity of unrelated drug classes if over-expressed. The first reports of drug resistant bacterial infections were reported in the 1940s after the first mass production of antibiotics. Most of the RND superfamily transport systems are made of large polypeptide chains. RND proteins exist primarily in gram-negative bacteria but can also be found in gram-positive bacteria, archaea, and eukaryotes.
Azohydromonas lata is a gram-negative, hydrogen-using bacterium from the genus Azohydromonas. Alcaligenes latus has been reclassified as Azohydromonas lata.











