
Cytochromes are redox-active proteins containing a heme, with a central iron (Fe) atom at its core, as a cofactor. They are involved in electron transport chain and redox catalysis. They are classified according to the type of heme and its mode of binding. Four varieties are recognized by the International Union of Biochemistry and Molecular Biology (IUBMB), cytochromes a, cytochromes b, cytochromes c and cytochrome d.

An electron transport chain (ETC) is a series of protein complexes and other molecules that transfer electrons from electron donors to electron acceptors via redox reactions (both reduction and oxidation occurring simultaneously) and couples this electron transfer with the transfer of protons (H+ ions) across a membrane. The electrons that are transferred from NADH and FADH2 to the ETC involves four multi-subunit large enzymes complexes and two mobile electron carriers. Many of the enzymes in the electron transport chain are embedded within the membrane.

The cytochrome complex, or cyt c, is a small hemeprotein found loosely associated with the inner membrane of the mitochondrion where it plays a critical role in cellular respiration. It transfers electrons between Complexes III and IV. Cytochrome c is highly water-soluble, unlike other cytochromes. It is capable of undergoing oxidation and reduction as its iron atom converts between the ferrous and ferric forms, but does not bind oxygen. It also plays a major role in cell apoptosis. In humans, cytochrome c is encoded by the CYCS gene.

A hemeprotein, or heme protein, is a protein that contains a heme prosthetic group. They are a very large class of metalloproteins. The heme group confers functionality, which can include oxygen carrying, oxygen reduction, electron transfer, and other processes. Heme is bound to the protein either covalently or noncovalently or both.

The enzyme cytochrome c oxidase or Complex IV, is a large transmembrane protein complex found in bacteria, archaea, and mitochondria of eukaryotes.

The coenzyme Q : cytochrome c – oxidoreductase, sometimes called the cytochrome bc1 complex, and at other times complex III, is the third complex in the electron transport chain, playing a critical role in biochemical generation of ATP. Complex III is a multisubunit transmembrane protein encoded by both the mitochondrial and the nuclear genomes. Complex III is present in the mitochondria of all animals and all aerobic eukaryotes and the inner membranes of most eubacteria. Mutations in Complex III cause exercise intolerance as well as multisystem disorders. The bc1 complex contains 11 subunits, 3 respiratory subunits, 2 core proteins and 6 low-molecular weight proteins.

Heme, or haem, is a precursor to hemoglobin, which is necessary to bind oxygen in the bloodstream. Heme is biosynthesized in both the bone marrow and the liver.

Cytochrome c peroxidase, or CCP, is a water-soluble heme-containing enzyme of the peroxidase family that takes reducing equivalents from cytochrome c and reduces hydrogen peroxide to water:

Succinate dehydrogenase (SDH) or succinate-coenzyme Q reductase (SQR) or respiratory complex II is an enzyme complex, found in many bacterial cells and in the inner mitochondrial membrane of eukaryotes. It is the only enzyme that participates in both the citric acid cycle and the electron transport chain. Histochemical analysis showing high succinate dehydrogenase in muscle demonstrates high mitochondrial content and high oxidative potential.

In biochemistry, flavin adenine dinucleotide (FAD) is a redox-active coenzyme associated with various proteins, which is involved with several enzymatic reactions in metabolism. A flavoprotein is a protein that contains a flavin group, which may be in the form of FAD or flavin mononucleotide (FMN). Many flavoproteins are known: components of the succinate dehydrogenase complex, α-ketoglutarate dehydrogenase, and a component of the pyruvate dehydrogenase complex.

Cytochrome f is the largest subunit of cytochrome b6f complex. In its structure and functions, the cytochrome b6f complex bears extensive analogy to the cytochrome bc1 complex of mitochondria and photosynthetic purple bacteria. Cytochrome f plays a role analogous to that of cytochrome c1, in spite of their different structures.

Rieske proteins are iron–sulfur protein (ISP) components of cytochrome bc1 complexes and cytochrome b6f complexes and are responsible for electron transfer in some biological systems. John S. Rieske and co-workers first discovered the protein and in 1964 isolated an acetylated form of the bovine mitochondrial protein. In 1979 Trumpower's lab isolated the "oxidation factor" from bovine mitochondria and showed it was a reconstitutively-active form of the Rieske iron-sulfur protein
It is a unique [2Fe-2S] cluster in that one of the two Fe atoms is coordinated by two histidine residues rather than two cysteine residues. They have since been found in plants, animals, and bacteria with widely ranging electron reduction potentials from -150 to +400 mV.

The inner mitochondrial membrane (IMM) is the mitochondrial membrane which separates the mitochondrial matrix from the intermembrane space.

Cytochromes b5 are ubiquitous electron transport hemoproteins found in animals, plants, fungi and purple phototrophic bacteria. The microsomal and mitochondrial variants are membrane-bound, while bacterial and those from erythrocytes and other animal tissues are water-soluble. The family of cytochrome b5-like proteins includes hemoprotein domains covalently associated with other redox domains in flavocytochrome cytochrome b2, sulfite oxidase, plant and fungal nitrate reductases, and plant and fungal cytochrome b5/acyl lipid desaturase fusion proteins.
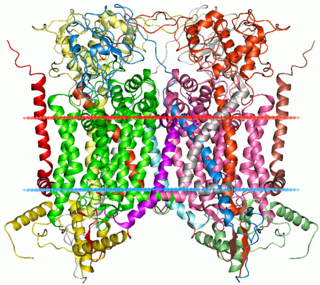
Cytochrome b within both molecular and cell biology, is a protein found in the mitochondria of eukaryotic cells. It functions as part of the electron transport chain and is the main subunit of transmembrane cytochrome bc1 and b6f complexes.

Heme C is an important kind of heme.
In enzymology, a cellobiose dehydrogenase (acceptor) (EC 1.1.99.18) is an enzyme that catalyzes the chemical reaction

The enzyme holocytochrome-c synthase catalyzes the chemical reaction
Haem peroxidases (or heme peroxidases) are haem-containing enzymes that use hydrogen peroxide as the electron acceptor to catalyse a number of oxidative reactions. Most haem peroxidases follow the reaction scheme:
In molecular biology, the cytochrome c assembly protein family includes various proteins involved in cytochrome c assembly from mitochondria and bacteria. Members of this family include: CycK from Rhizobium leguminosarum, CcmC from Escherichia coli and Paracoccus denitrificans, and orf240 from Triticum aestivum (Wheat) mitochondria. The members of this family are probably integral membrane proteins with six predicted transmembrane helices that may comprise the membrane component of an ABC transporter complex. This transporter may be necessary for transport of some component needed for cytochrome c assembly. One member, R. leguminosarum CycK, contains a putative haem-binding motif. Wheat orf240 also contains a putative haem-binding motif and is a proposed ABC transporter with c-type haem as its proposed substrate. However it seems unlikely that all members of this family transport haem or c-type apocytochromes because P. denitrificans CcmC transports neither.



















