Related Research Articles

The G0 phase describes a cellular state outside of the replicative cell cycle. Classically, cells were thought to enter G0 primarily due to environmental factors, like nutrient deprivation, that limited the resources necessary for proliferation. Thus it was thought of as a resting phase. G0 is now known to take different forms and occur for multiple reasons. For example, most adult neuronal cells, among the most metabolically active cells in the body, are fully differentiated and reside in a terminal G0 phase. Neurons reside in this state, not because of stochastic or limited nutrient supply, but as a part of their developmental program.

In chemistry, reactive oxygen species (ROS) are highly reactive chemicals formed from diatomic oxygen. Examples of ROS include peroxides, superoxide, hydroxyl radical, singlet oxygen, and alpha-oxygen.

DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA damage, resulting in tens of thousands of individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur, including double-strand breaks and DNA crosslinkages. This can eventually lead to malignant tumors, or cancer as per the two hit hypothesis.

Fibrosis, also known as fibrotic scarring, is a pathological wound healing in which connective tissue replaces normal parenchymal tissue to the extent that it goes unchecked, leading to considerable tissue remodelling and the formation of permanent scar tissue.

Poly (ADP-ribose) polymerase (PARP) is a family of proteins involved in a number of cellular processes such as DNA repair, genomic stability, and programmed cell death.

p16, is a protein that slows cell division by slowing the progression of the cell cycle from the G1 phase to the S phase, thereby acting as a tumor suppressor. It is encoded by the CDKN2A gene. A deletion in this gene can result in insufficient or non-functional p16, accelerating the cell cycle and resulting in many types of cancer.

H2A histone family member X is a type of histone protein from the H2A family encoded by the H2AFX gene. An important phosphorylated form is γH2AX (S139), which forms when double-strand breaks appear.
Chromatin remodeling is the dynamic modification of chromatin architecture to allow access of condensed genomic DNA to the regulatory transcription machinery proteins, and thereby control gene expression. Such remodeling is principally carried out by 1) covalent histone modifications by specific enzymes, e.g., histone acetyltransferases (HATs), deacetylases, methyltransferases, and kinases, and 2) ATP-dependent chromatin remodeling complexes which either move, eject or restructure nucleosomes. Besides actively regulating gene expression, dynamic remodeling of chromatin imparts an epigenetic regulatory role in several key biological processes, egg cells DNA replication and repair; apoptosis; chromosome segregation as well as development and pluripotency. Aberrations in chromatin remodeling proteins are found to be associated with human diseases, including cancer. Targeting chromatin remodeling pathways is currently evolving as a major therapeutic strategy in the treatment of several cancers.
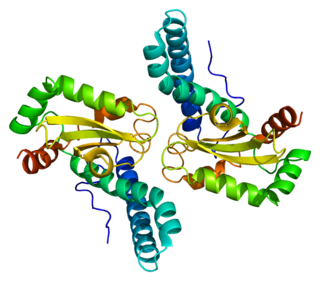
Superoxide dismutase 2, mitochondrial (SOD2), also known as manganese-dependent superoxide dismutase (MnSOD), is an enzyme which in humans is encoded by the SOD2 gene on chromosome 6. A related pseudogene has been identified on chromosome 1. Alternative splicing of this gene results in multiple transcript variants. This gene is a member of the iron/manganese superoxide dismutase family. It encodes a mitochondrial protein that forms a homotetramer and binds one manganese ion per subunit. This protein binds to the superoxide byproducts of oxidative phosphorylation and converts them to hydrogen peroxide and diatomic oxygen. Mutations in this gene have been associated with idiopathic cardiomyopathy (IDC), premature aging, sporadic motor neuron disease, and cancer.

Promyelocytic leukemia protein (PML) is the protein product of the PML gene. PML protein is a tumor suppressor protein required for the assembly of a number of nuclear structures, called PML-nuclear bodies, which form amongst the chromatin of the cell nucleus. These nuclear bodies are present in mammalian nuclei, at about 1 to 30 per cell nucleus. PML-NBs are known to have a number of regulatory cellular functions, including involvement in programmed cell death, genome stability, antiviral effects and controlling cell division. PML mutation or loss, and the subsequent dysregulation of these processes, has been implicated in a variety of cancers.

High-mobility group protein B2 also known as high-mobility group protein 2 (HMG-2) is a protein that in humans is encoded by the HMGB2 gene.
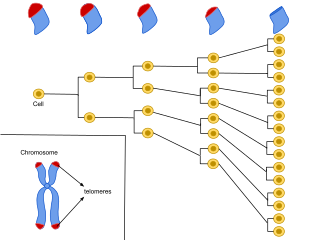
Cellular senescence is a phenomenon characterized by the cessation of cell division. In their experiments during the early 1960s, Leonard Hayflick and Paul Moorhead found that normal human fetal fibroblasts in culture reach a maximum of approximately 50 cell population doublings before becoming senescent. This process is known as "replicative senescence", or the Hayflick limit. Hayflick's discovery of mortal cells paved the path for the discovery and understanding of cellular aging molecular pathways. Cellular senescence can be initiated by a wide variety of stress inducing factors. These stress factors include both environmental and internal damaging events, abnormal cellular growth, oxidative stress, autophagy factors, among many other things.

Yippee-like 3 (Drosophila) is a protein that in humans is encoded by the YPEL3 gene. YPEL3 has growth inhibitory effects in normal and tumor cell lines. One of five family members (YPEL1-5), YPEL3 was named in reference to its Drosophila melanogaster orthologue. Initially discovered in a gene expression profiling assay of p53 activated MCF7 cells, induction of YPEL3 has been shown to trigger permanent growth arrest or cellular senescence in certain human normal and tumor cell types. DNA methylation of a CpG island near the YPEL3 promoter as well as histone acetylation may represent possible epigenetic mechanisms leading to decreased gene expression in human tumors.

The retinoblastoma protein is a tumor suppressor protein that is dysfunctional in several major cancers. One function of pRb is to prevent excessive cell growth by inhibiting cell cycle progression until a cell is ready to divide. When the cell is ready to divide, pRb is phosphorylated, inactivating it, and the cell cycle is allowed to progress. It is also a recruiter of several chromatin remodeling enzymes such as methylases and acetylases.
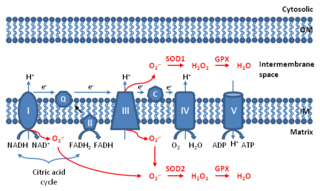
Mitochondrial ROS are reactive oxygen species (ROS) that are produced by mitochondria. Generation of mitochondrial ROS mainly takes place at the electron transport chain located on the inner mitochondrial membrane during the process of oxidative phosphorylation. Leakage of electrons at complex I and complex III from electron transport chains leads to partial reduction of oxygen to form superoxide. Subsequently, superoxide is quickly dismutated to hydrogen peroxide by two dismutases including superoxide dismutase 2 (SOD2) in mitochondrial matrix and superoxide dismutase 1 (SOD1) in mitochondrial intermembrane space. Collectively, both superoxide and hydrogen peroxide generated in this process are considered as mitochondrial ROS.
Androgen deprivation-induced senescence refers to the induction of cellular senescence as a result of androgen deprivation therapy. ADIS is observed in prostate cancer cells that are dependent on androgens for cell proliferation. Androgen withdrawal induces cells to undergo cellular senescence by up-regulating intracellular reactive oxygen species (ROS) that cause DNA damage. ADIS is maintained through the up-regulation of the cell cycle regulator p16ink4a.
Judith Campisi is an American biochemist and cell biologist. She is a professor of biogerontology at the Buck Institute for Research on Aging. She is also a member of the SENS Research Foundation Advisory Board and an adviser at the Lifeboat Foundation. She is co-editor in chief of the Aging Journal, together with Mikhail Blagosklonny and David Sinclair, and founder of the pharmaceutical company Unity Biotechnology. She is listed in Who's Who in Gerontology. She is widely known for her research on how senescent cells influence aging and cancer — in particular the Senescence Associated Secretory Phenotype (SASP).
Senotherapy is an early-stage basic research field for development of possible therapeutic agents and strategies to specifically target cellular senescence, an altered cell state associated with ageing and age-related diseases. The name derives from intent of the proposed anti-aging drug to halt "senescence". As of 2019, much of the research remains preliminary and there are no drugs approved for this purpose.
Senescence-associated secretory phenotype (SASP) is a phenotype associated with senescent cells wherein those cells secrete high levels of inflammatory cytokines, immune modulators, growth factors, and proteases. SASP may also consist of exosomes and ectosomes containing enzymes, microRNA, DNA fragments, chemokines, and other bioactive factors. Soluble urokinase plasminogen activator surface receptor is part of SASP, and has been used to identify senescent cells for senolytic therapy. Initially, SASP is immunosuppressive and profibrotic, but progresses to become proinflammatory and fibrolytic. SASP is the primary cause of the detrimental effects of senescent cells.
The hallmarks of aging are the types of biochemical changes that occur in all organisms that experience biological aging and lead to a progressive loss of physiological integrity, impaired function and, eventually, death. They were first listed in a landmark paper in 2013 to conceptualize the essence of biological aging and its underlying mechanisms.
References
- 1 2 3 Rodier, F; Muñoz, DP; Teachenor, R; Chu, V; Le, O; Bhaumik, D; Coppé, JP; Campeau, E; Beauséjour, CM; Kim, SH; Davalos, AR; Campisi, J (1 January 2011). "DNA-SCARS: distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion". Journal of Cell Science. 124 (Pt 1): 68–81. doi:10.1242/jcs.071340. PMC 3001408 . PMID 21118958.
- ↑ Rodier, F.; Campisi, J. (14 February 2011). "Four faces of cellular senescence". The Journal of Cell Biology. 192 (4): 547–556. doi:10.1083/jcb.201009094. PMC 3044123 . PMID 21321098.