
Tumor necrosis factor (TNF), formerly known as TNF-α, is an inflammatory protein and a principal mediator of the innate immune response. TNF is produced primarily by macrophages in response to antigens, and activates inflammatory pathways through its two receptors, TNFR1 and TNFR2. It is a member of the tumor necrosis factor superfamily, a family of type II transmembrane proteins that function as cytokines. Excess production of TNF plays a critical role in the pathology of several inflammatory diseases, and anti-TNF therapies are often employed to treat these diseases.

Pyridoxal phosphate (PLP, pyridoxal 5'-phosphate, P5P), the active form of vitamin B6, is a coenzyme in a variety of enzymatic reactions. The International Union of Biochemistry and Molecular Biology has catalogued more than 140 PLP-dependent activities, corresponding to ~4% of all classified activities. The versatility of PLP arises from its ability to covalently bind the substrate, and then to act as an electrophilic catalyst, thereby stabilizing different types of carbanionic reaction intermediates.
Biosynthesis, i.e., chemical synthesis occurring in biological contexts, is a term most often referring to multi-step, enzyme-catalyzed processes where chemical substances absorbed as nutrients serve as enzyme substrates, with conversion by the living organism either into simpler or more complex products. Examples of biosynthetic pathways include those for the production of amino acids, lipid membrane components, and nucleotides, but also for the production of all classes of biological macromolecules, and of acetyl-coenzyme A, adenosine triphosphate, nicotinamide adenine dinucleotide and other key intermediate and transactional molecules needed for metabolism. Thus, in biosynthesis, any of an array of compounds, from simple to complex, are converted into other compounds, and so it includes both the catabolism and anabolism of complex molecules. Biosynthetic processes are often represented via charts of metabolic pathways. A particular biosynthetic pathway may be located within a single cellular organelle, while others involve enzymes that are located across an array of cellular organelles and structures.

Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) is a family of transcription factor protein complexes that controls transcription of DNA, cytokine production and cell survival. NF-κB is found in almost all animal cell types and is involved in cellular responses to stimuli such as stress, cytokines, free radicals, heavy metals, ultraviolet irradiation, oxidized LDL, and bacterial or viral antigens. NF-κB plays a key role in regulating the immune response to infection. Incorrect regulation of NF-κB has been linked to cancer, inflammatory and autoimmune diseases, septic shock, viral infection, and improper immune development. NF-κB has also been implicated in processes of synaptic plasticity and memory.

Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding S-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltransferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the leaving group and the methyl group attached to it acts as the electrophile that transfers the methyl group to the enzyme substrate. SAM is converted to S-Adenosyl homocysteine (SAH) during this process. The breaking of the SAM-methyl bond and the formation of the substrate-methyl bond happen nearly simultaneously. These enzymatic reactions are found in many pathways and are implicated in genetic diseases, cancer, and metabolic diseases. Another type of methyl transfer is the radical S-Adenosyl methionine (SAM) which is the methylation of unactivated carbon atoms in primary metabolites, proteins, lipids, and RNA.

Chemogenomics, or chemical genomics, is the systematic screening of targeted chemical libraries of small molecules against individual drug target families with the ultimate goal of identification of novel drugs and drug targets. Typically some members of a target library have been well characterized where both the function has been determined and compounds that modulate the function of those targets have been identified. Other members of the target family may have unknown function with no known ligands and hence are classified as orphan receptors. By identifying screening hits that modulate the activity of the less well characterized members of the target family, the function of these novel targets can be elucidated. Furthermore, the hits for these targets can be used as a starting point for drug discovery. The completion of the human genome project has provided an abundance of potential targets for therapeutic intervention. Chemogenomics strives to study the intersection of all possible drugs on all of these potential targets.
Diphtheria toxin is an exotoxin secreted mainly by Corynebacterium diphtheriae but also by Corynebacterium ulcerans and Corynebacterium pseudotuberculosis, the pathogenic bacterium that causes diphtheria. The toxin gene is encoded by a prophage called corynephage β. The toxin causes the disease in humans by gaining entry into the cell cytoplasm and inhibiting protein synthesis.

Elongation factors are a set of proteins that function at the ribosome, during protein synthesis, to facilitate translational elongation from the formation of the first to the last peptide bond of a growing polypeptide. Most common elongation factors in prokaryotes are EF-Tu, EF-Ts, EF-G. Bacteria and eukaryotes use elongation factors that are largely homologous to each other, but with distinct structures and different research nomenclatures.
Anthrax toxin is a three-protein exotoxin secreted by virulent strains of the bacterium, Bacillus anthracis—the causative agent of anthrax. The toxin was first discovered by Harry Smith in 1954. Anthrax toxin is composed of a cell-binding protein, known as protective antigen (PA), and two enzyme components, called edema factor (EF) and lethal factor (LF). These three protein components act together to impart their physiological effects. Assembled complexes containing the toxin components are endocytosed. In the endosome, the enzymatic components of the toxin translocate into the cytoplasm of a target cell. Once in the cytosol, the enzymatic components of the toxin disrupt various immune cell functions, namely cellular signaling and cell migration. The toxin may even induce cell lysis, as is observed for macrophage cells. Anthrax toxin allows the bacteria to evade the immune system, proliferate, and ultimately kill the host animal. Research on anthrax toxin also provides insight into the generation of macromolecular assemblies, and on protein translocation, pore formation, endocytosis, and other biochemical processes.

Amino acid biosynthesis is the set of biochemical processes by which the amino acids are produced. The substrates for these processes are various compounds in the organism's diet or growth media. Not all organisms are able to synthesize all amino acids. For example, humans can synthesize 11 of the 20 standard amino acids. These 11 are called the non-essential amino acids.

Gliotoxin is a sulfur-containing mycotoxin that belongs to a class of naturally occurring 2,5-diketopiperazines produced by several species of fungi, especially those of marine origin. It is the most prominent member of the epipolythiopiperazines, a large class of natural products featuring a diketopiperazine with di- or polysulfide linkage. These highly bioactive compounds have been the subject of numerous studies aimed at new therapeutics. Gliotoxin was originally isolated from Gliocladium fimbriatum, and was named accordingly. It is an epipolythiodioxopiperazine metabolite that is one of the most abundantly produced metabolites in human invasive Aspergillosis (IA).
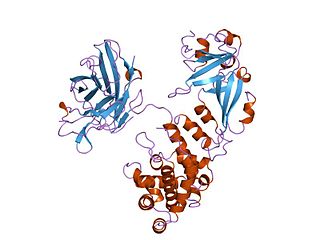
IKK-β also known as inhibitor of nuclear factor kappa-B kinase subunit beta is a protein that in humans is encoded by the IKBKB gene.
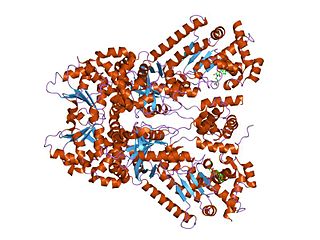
Transcription factor RelB is a protein that in humans is encoded by the RELB gene.

Eukaryotic elongation factor 2 is a protein that in humans is encoded by the EEF2 gene. It is the archaeal and eukaryotic counterpart of bacterial EF-G.

Diphthine synthase is an enzyme that in humans is encoded by the DPH5 gene.

Diphthamide biosynthesis protein 1 is a protein that in humans is encoded by the DPH1 gene. It encodes a protein that performs posttranslational modification of histidine-715 on eukaryotic translation elongation factor 2 to diphthamide. This modification appears to be important in the translation of Cyclin D in ovarian cells. DPH1 is mutated in 90% of ovarian cancers end stage, usually by loss of heterozygosity.

Withaferin A is a steroidal lactone, derived from Acnistus arborescens, Withania somnifera and other members of family Solanaceae. It is the first member of the withanolide class of ergostane type product to be discovered.
Radical SAM enzymes belong to a superfamily of enzymes that use an iron-sulfur cluster (4Fe-4S) to reductively cleave S-adenosyl-L-methionine (SAM) to generate a radical, usually a 5′-deoxyadenosyl radical (5'-dAdo), as a critical intermediate. These enzymes utilize this radical intermediate to perform diverse transformations, often to functionalize unactivated C-H bonds. Radical SAM enzymes are involved in cofactor biosynthesis, enzyme activation, peptide modification, post-transcriptional and post-translational modifications, metalloprotein cluster formation, tRNA modification, lipid metabolism, biosynthesis of antibiotics and natural products etc. The vast majority of known radical SAM enzymes belong to the radical SAM superfamily, and have a cysteine-rich motif that matches or resembles CxxxCxxC. Radical SAM enzymes comprise the largest superfamily of metal-containing enzymes.
Curcumin synthase categorizes three enzyme isoforms, type III polyketide synthases (PKSs) present in the leaves and rhizome of the turmeric plant that synthesize curcumin. CURS1-3 are responsible for the hydrolysis of feruloyldiketide-CoA, previously produced in the curcuminoid pathway, and a decarboxylative condensation reaction that together comprise one of the final steps in the synthesis pathway for curcumin, demethoxycurcumin, and bisdemethoxycurcumin, the compounds that give turmeric both its distinctive yellow color, and traditional medical benefits. CURS should not be confused with Curcuminoid Synthase (CUS), which catalyzes the one-pot synthesis of bisdemethoxycurcumin in Oryza sativa.
The interleukin-1 receptor (IL-1R) associated kinase (IRAK) family plays a crucial role in the protective response to pathogens introduced into the human body by inducing acute inflammation followed by additional adaptive immune responses. IRAKs are essential components of the Interleukin-1 receptor signaling pathway and some Toll-like receptor signaling pathways. Toll-like receptors (TLRs) detect microorganisms by recognizing specific pathogen-associated molecular patterns (PAMPs) and IL-1R family members respond the interleukin-1 (IL-1) family cytokines. These receptors initiate an intracellular signaling cascade through adaptor proteins, primarily, MyD88. This is followed by the activation of IRAKs. TLRs and IL-1R members have a highly conserved amino acid sequence in their cytoplasmic domain called the Toll/Interleukin-1 (TIR) domain. The elicitation of different TLRs/IL-1Rs results in similar signaling cascades due to their homologous TIR motif leading to the activation of mitogen-activated protein kinases (MAPKs) and the IκB kinase (IKK) complex, which initiates a nuclear factor-κB (NF-κB) and AP-1-dependent transcriptional response of pro-inflammatory genes. Understanding the key players and their roles in the TLR/IL-1R pathway is important because the presence of mutations causing the abnormal regulation of Toll/IL-1R signaling leading to a variety of acute inflammatory and autoimmune diseases.
















