Parkin is a 465-amino acid residue E3 ubiquitin ligase, a protein that in humans and mice is encoded by the PARK2 gene. Parkin plays a critical role in ubiquitination – the process whereby molecules are covalently labelled with ubiquitin (Ub) and directed towards degradation in proteasomes or lysosomes. Ubiquitination involves the sequential action of three enzymes. First, an E1 ubiquitin-activating enzyme binds to inactive Ub in eukaryotic cells via a thioester bond and mobilises it in an ATP-dependent process. Ub is then transferred to an E2 ubiquitin-conjugating enzyme before being conjugated to the target protein via an E3 ubiquitin ligase. There exists a multitude of E3 ligases, which differ in structure and substrate specificity to allow selective targeting of proteins to intracellular degradation.

Leber's hereditary optic neuropathy (LHON) is a mitochondrially inherited degeneration of retinal ganglion cells (RGCs) and their axons that leads to an acute or subacute loss of central vision; it predominantly affects young adult males. LHON is transmitted only through the mother, as it is primarily due to mutations in the mitochondrial genome, and only the egg contributes mitochondria to the embryo. Men cannot pass on the disease to their offspring. LHON is usually due to one of three pathogenic mitochondrial DNA (mtDNA) point mutations. These mutations are at nucleotide positions 11778 G to A, 3460 G to A and 14484 T to C, respectively in the ND4, ND1 and ND6 subunit genes of complex I of the oxidative phosphorylation chain in mitochondria.

Behr syndrome is characterized by the association of early-onset optic atrophy with spinocerebellar degeneration resulting in ataxia, pyramidal signs, peripheral neuropathy and developmental delay.
Dominant optic atrophy (DOA), or autosomal dominant optic atrophy (ADOA), (Kjer's type) is an autosomally inherited disease that affects the optic nerves, causing reduced visual acuity and blindness beginning in childhood. However, the disease can seem to re-present a second time with further vision loss due to the early onset of presbyopia symptoms (i.e., difficulty in viewing objects up close). DOA is characterized as affecting neurons called retinal ganglion cells (RGCs). This condition is due to mitochondrial dysfunction mediating the death of optic nerve fibers. The RGCs axons form the optic nerve. Therefore, the disease can be considered of the central nervous system. Dominant optic atrophy was first described clinically by Batten in 1896 and named Kjer’s optic neuropathy in 1959 after Danish ophthalmologist Poul Kjer, who studied 19 families with the disease. Although dominant optic atrophy is the most common autosomally inherited optic neuropathy (i.e., disease of the optic nerves), it is often misdiagnosed.
Dynamin is a GTPase responsible for endocytosis in the eukaryotic cell. Dynamin is part of the "dynamin superfamily", which includes classical dynamins, dynamin-like proteins, Mx proteins, OPA1, mitofusins, and GBPs. Members of the dynamin family are principally involved in the scission of newly formed vesicles from the membrane of one cellular compartment and their targeting to, and fusion with, another compartment, both at the cell surface as well as at the Golgi apparatus. Dynamin family members also play a role in many processes including division of organelles, cytokinesis and microbial pathogen resistance.

Costeff syndrome, or 3-methylglutaconic aciduria type III, is a genetic disorder caused by mutations in the OPA3 gene. It is typically associated with the onset of visual deterioration in early childhood followed by the development of movement problems and motor disability in later childhood, occasionally along with mild cases of cognitive deficiency. The disorder is named after Hanan Costeff, the doctor who first described the syndrome in 1989.

Mitofusin-2 is a protein that in humans is encoded by the MFN2 gene. Mitofusins are GTPases embedded in the outer membrane of the mitochondria. In mammals MFN1 and MFN2 are essential for mitochondrial fusion. In addition to the mitofusins, OPA1 regulates inner mitochondrial membrane fusion, and DRP1 is responsible for mitochondrial fission.
Dynamin-2 is a protein that in humans is encoded by the DNM2 gene.

X-linked retinitis pigmentosa GTPase regulator is a GTPase-binding protein that in humans is encoded by the RPGR gene. The gene is located on the X-chromosome and is commonly associated with X-linked retinitis pigmentosa (XLRP). In photoreceptor cells, RPGR is localized in the connecting cilium which connects the protein-synthesizing inner segment to the photosensitive outer segment and is involved in the modulation of cargo trafficked between the two segments.

Mitochondrial import inner membrane translocase subunit Tim8 A, also known as deafness-dystonia peptide or protein is an enzyme that in humans is encoded by the TIMM8A gene. This translocase has similarity to yeast mitochondrial proteins that are involved in the import of metabolite transporters from the cytoplasm into the mitochondrial inner membrane. The gene is mutated in deafness-dystonia syndrome and it is postulated that MTS/DFN-1 is a mitochondrial disease caused by a defective mitochondrial protein import system.

Dynamin-1-like protein is a GTPase that regulates mitochondrial fission. In humans, dynamin-1-like protein, which is typically referred to as dynamin-related protein 1 (Drp1), is encoded by the DNM1L gene and is part of the dynamin superfamily (DSP) family of proteins.

ATP-dependent metalloprotease YME1L1 is an enzyme that in humans is encoded by the YME1L1 gene. YME1L1 belongs to the AAA family of ATPases and mainly functions in the maintenance of mitochondrial morphology. Mutations in this gene would cause infantile-onset mitochondriopathy.

Presenilins-associated rhomboid-like protein, mitochondrial (PSARL), also known as PINK1/PGAM5-associated rhomboid-like protease (PARL), is an inner mitochondrial membrane protein that in humans is encoded by the PARL gene on chromosome 3. It is a member of the rhomboid family of intramembrane serine proteases. This protein is involved in signal transduction and apoptosis, as well as neurodegenerative diseases and type 2 diabetes.

Caseinolytic peptidase B protein homolog (CLPB), also known as Skd3, is a mitochondrial AAA ATPase chaperone that in humans is encoded by the gene CLPB, which encodes an adenosine triphosphate-(ATP) dependent chaperone. Skd3 is localized in mitochondria and widely expressed in human tissues. High expression in adult brain and low expression in granulocyte is found. It is a potent protein disaggregase that chaperones the mitochondrial intermembrane space. Mutations in the CLPB gene could cause autosomal recessive metabolic disorder with intellectual disability/developmental delay, congenital neutropenia, progressive brain atrophy, movement disorder, cataracts, and 3-methylglutaconic aciduria. Recently, heterozygous, dominant negative mutations in CLPB have been identified as a cause of severe congenital neutropenia (SCN).
Paul Kjer is a Danish ophthalmologist who studied a condition in nineteen families that was characterized by infantile optic atrophy along with a dominant inheritance mode. In 1959, the condition was named Kjer's optic neuropathy in his honor.
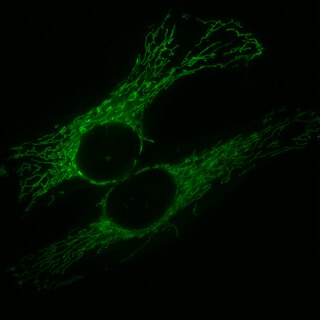
Mitochondria are dynamic organelles with the ability to fuse and divide (fission), forming constantly changing tubular networks in most eukaryotic cells. These mitochondrial dynamics, first observed over a hundred years ago are important for the health of the cell, and defects in dynamics lead to genetic disorders. Through fusion, mitochondria can overcome the dangerous consequences of genetic malfunction. The process of mitochondrial fusion involves a variety of proteins that assist the cell throughout the series of events that form this process.
Luca Scorrano is an Italian biologist and professor of Biochemistry at the University of Padua as well as the former Scientific Director of the Veneto Institute of Molecular Medicine in Italy. He is known for his important contributions to the field of mitochondrial dynamics and the interface between mitochondria and the endoplasmic reticulum.
Coiled-coil-helix-coiled-coil-helix domain-containing protein 10, mitochondrial, also known as Protein N27C7-4 is a protein that in humans is encoded by the CHCHD10 gene.

Solute carrier family 25 member 46 is a protein that in humans is encoded by the SLC25A46 gene. This protein is a member of the SLC25 mitochondrial solute carrier family. It is a transmembrane protein located in the mitochondrial outer membrane involved in lipid transfer from the endoplasmic reticulum (ER) to mitochondria. Mutations in this gene result in neuropathy and optic atrophy.
Taosheng Huang is a physician-scientist with substantial academic achievements and professional experience in translational research, specifically, in human mitochondrial genetics. He is a full Professor and Director of the Molecular Diagnostic Laboratory in the Division of Human Genetics at Cincinnati Children’s Hospital Medical Center (CCHMC). Huang has published over 100 manuscripts in many impactful journals.