A restriction enzyme, restriction endonuclease, or restrictase is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone of the DNA double helix.

In biochemistry, allosteric regulation is the regulation of an enzyme by binding an effector molecule at a site other than the enzyme's active site.

In biochemistry, the DNA methyltransferase family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl methionine (SAM) as the methyl donor.
In cell biology, protein kinase A (PKA) is a family of enzymes whose activity is dependent on cellular levels of cyclic AMP (cAMP). PKA is also known as cAMP-dependent protein kinase. PKA has several functions in the cell, including regulation of glycogen, sugar, and lipid metabolism.
The restriction modification system is found in bacteria and other prokaryotic organisms, and provides a defense against foreign DNA, such as that borne by bacteriophages.

Aspartate carbamoyltransferase catalyzes the first step in the pyrimidine biosynthetic pathway.

Ribonuclease H is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes.

RecBCD is an enzyme of the E. coli bacterium that initiates recombinational repair from potentially lethal double strand breaks in DNA which may result from ionizing radiation, replication errors, endonucleases, oxidative damage, and a host of other factors. The RecBCD enzyme is both a helicase that unwinds, or separates the strands of DNA, and a nuclease that makes single-stranded nicks in DNA.

Dihydrolipoyl transacetylase is an enzyme component of the multienzyme pyruvate dehydrogenase complex. The pyruvate dehydrogenase complex is responsible for the pyruvate decarboxylation step that links glycolysis to the citric acid cycle. This involves the transformation of pyruvate from glycolysis into acetyl-CoA which is then used in the citric acid cycle to carry out cellular respiration.
Tyrosine hydroxylase or tyrosine 3-monooxygenase is the enzyme responsible for catalyzing the conversion of the amino acid L-tyrosine to L-3,4-dihydroxyphenylalanine (L-DOPA). It does so using molecular oxygen (O2), as well as iron (Fe2+) and tetrahydrobiopterin as cofactors. L-DOPA is a precursor for dopamine, which, in turn, is a precursor for the important neurotransmitters norepinephrine (noradrenaline) and epinephrine (adrenaline). Tyrosine hydroxylase catalyzes the rate limiting step in this synthesis of catecholamines. In humans, tyrosine hydroxylase is encoded by the TH gene, and the enzyme is present in the central nervous system (CNS), peripheral sympathetic neurons and the adrenal medulla. Tyrosine hydroxylase, phenylalanine hydroxylase and tryptophan hydroxylase together make up the family of aromatic amino acid hydroxylases (AAAHs).

A DNA clamp, also known as a sliding clamp or β-clamp, is a protein complex that serves as a processivity-promoting factor in DNA replication. As a critical component of the DNA polymerase III holoenzyme, the clamp protein binds DNA polymerase and prevents this enzyme from dissociating from the template DNA strand. The clamp-polymerase protein–protein interactions are stronger and more specific than the direct interactions between the polymerase and the template DNA strand; because one of the rate-limiting steps in the DNA synthesis reaction is the association of the polymerase with the DNA template, the presence of the sliding clamp dramatically increases the number of nucleotides that the polymerase can add to the growing strand per association event. The presence of the DNA clamp can increase the rate of DNA synthesis up to 1,000-fold compared with a nonprocessive polymerase.

Phosphorylase kinase (PhK) is a serine/threonine-specific protein kinase which activates glycogen phosphorylase to release glucose-1-phosphate from glycogen. PhK phosphorylates glycogen phosphorylase at two serine residues, triggering a conformational shift which favors the more active glycogen phosphorylase “a” form over the less active glycogen phosphorylase b.
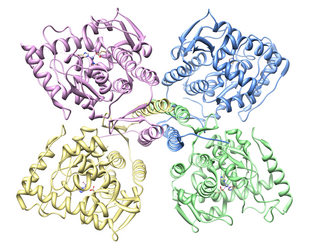
HindIII (pronounced "Hin D Three") is a type II site-specific deoxyribonuclease restriction enzyme isolated from Haemophilus influenzae that cleaves the DNA palindromic sequence AAGCTT in the presence of the cofactor Mg2+ via hydrolysis.

The homing endonucleases are a collection of endonucleases encoded either as freestanding genes within introns, as fusions with host proteins, or as self-splicing inteins. They catalyze the hydrolysis of genomic DNA within the cells that synthesize them, but do so at very few, or even singular, locations. Repair of the hydrolyzed DNA by the host cell frequently results in the gene encoding the homing endonuclease having been copied into the cleavage site, hence the term 'homing' to describe the movement of these genes. Homing endonucleases can thereby transmit their genes horizontally within a host population, increasing their allele frequency at greater than Mendelian rates.

Complement component 1 Q subcomponent-binding protein, mitochondrial is a protein that in humans is encoded by the C1QBP gene.
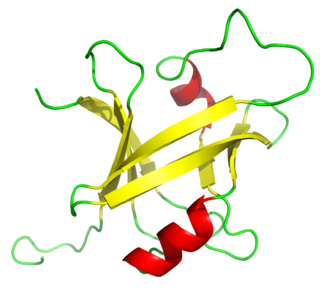
The B3 DNA binding domain (DBD) is a highly conserved domain found exclusively in transcription factors combined with other domains. It consists of 100-120 residues, includes seven beta strands and two alpha helices that form a DNA-binding pseudobarrel protein fold ; it interacts with the major groove of DNA.
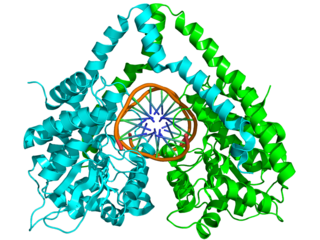
EcoRI is a restriction endonuclease enzyme isolated from species E. coli. It is a restriction enzyme that cleaves DNA double helices into fragments at specific sites, and is also a part of the restriction modification system. The Eco part of the enzyme's name originates from the species from which it was isolated - "E" denotes generic name which is "Escherichia" and "co" denotes species name, "coli" - while the R represents the particular strain, in this case RY13, and the I denotes that it was the first enzyme isolated from this strain.













