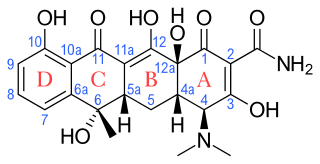An antibiotic is a type of antimicrobial substance active against bacteria. It is the most important type of antibacterial agent for fighting bacterial infections, and antibiotic medications are widely used in the treatment and prevention of such infections. They may either kill or inhibit the growth of bacteria. A limited number of antibiotics also possess antiprotozoal activity. Antibiotics are not effective against viruses such as the common cold or influenza; drugs which inhibit viruses are termed antiviral drugs or antivirals rather than antibiotics.

Antimicrobial resistance (AMR) occurs when microbes evolve mechanisms that protect them from the effects of antimicrobials. Antibiotic resistance is a subset of AMR, that applies specifically to bacteria that become resistant to antibiotics.

Staphylococcus aureus is a Gram-positive round-shaped bacterium, a member of the Firmicutes, and is a usual member of the microbiota of the body, frequently found in the upper respiratory tract and on the skin. It is often positive for catalase and nitrate reduction and is a facultative anaerobe that can grow without the need for oxygen. Although S. aureus usually acts as a commensal of the human microbiota it can also become an opportunistic pathogen, being a common cause of skin infections including abscesses, respiratory infections such as sinusitis, and food poisoning. Pathogenic strains often promote infections by producing virulence factors such as potent protein toxins, and the expression of a cell-surface protein that binds and inactivates antibodies. S. aureus is one of the leading pathogens for deaths associated with Antimicrobial resistance and the emergence of antibiotic-resistant strains such as methicillin-resistant S. aureus (MRSA) is a worldwide problem in clinical medicine. Despite much research and development, no vaccine for S. aureus has been approved.

Drug resistance is the reduction in effectiveness of a medication such as an antimicrobial or an antineoplastic in treating a disease or condition. The term is used in the context of resistance that pathogens or cancers have "acquired", that is, resistance has evolved. Antimicrobial resistance and antineoplastic resistance challenge clinical care and drive research. When an organism is resistant to more than one drug, it is said to be multidrug-resistant.

The Actinomycetota are a phylum of mostly Gram-positive bacteria. They can be terrestrial or aquatic. They are of great economic importance to humans because agriculture and forests depend on their contributions to soil systems. In soil they help to decompose the organic matter of dead organisms so the molecules can be taken up anew by plants. While this role is also played by fungi, Actinomycetota are much smaller and likely do not occupy the same ecological niche. In this role the colonies often grow extensive mycelia, like a fungus would, and the name of an important order of the phylum, Actinomycetales, reflects that they were long believed to be fungi. Some soil actinomycetota live symbiotically with the plants whose roots pervade the soil, fixing nitrogen for the plants in exchange for access to some of the plant's saccharides. Other species, such as many members of the genus Mycobacterium, are important pathogens.

Metagenomics is the study of genetic material recovered directly from environmental samples. The broad field may also be referred to as environmental genomics, ecogenomics or community genomics.

Antimicrobial peptides (AMPs), also called host defence peptides (HDPs) are part of the innate immune response found among all classes of life. Fundamental differences exist between prokaryotic and eukaryotic cells that may represent targets for antimicrobial peptides. These peptides are potent, broad spectrum antibiotics which demonstrate potential as novel therapeutic agents. Antimicrobial peptides have been demonstrated to kill Gram negative and Gram positive bacteria, enveloped viruses, fungi and even transformed or cancerous cells. Unlike the majority of conventional antibiotics it appears that antimicrobial peptides frequently destabilize biological membranes, can form transmembrane channels, and may also have the ability to enhance immunity by functioning as immunomodulators.
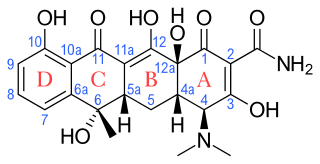
Tetracyclines are a group of broad-spectrum antibiotic compounds that have a common basic structure and are either isolated directly from several species of Streptomyces bacteria or produced semi-synthetically from those isolated compounds. Tetracycline molecules comprise a linear fused tetracyclic nucleus to which a variety of functional groups are attached. Tetracyclines are named for their four ("tetra-") hydrocarbon rings ("-cycl-") derivation ("-ine"). They are defined as a subclass of polyketides, having an octahydrotetracene-2-carboxamide skeleton and are known as derivatives of polycyclic naphthacene carboxamide. While all tetracyclines have a common structure, they differ from each other by the presence of chloride, methyl, and hydroxyl groups. These modifications do not change their broad antibacterial activity, but do affect pharmacological properties such as half-life and binding to proteins in serum.
In microbiology, the minimum inhibitory concentration (MIC) is the lowest concentration of a chemical, usually a drug, which prevents visible growth of a bacterium or bacteria. MIC depends on the microorganism, the affected human being, and the antibiotic itself. It is often expressed in micrograms per milliliter (μg/mL) or milligrams per liter (mg/L).
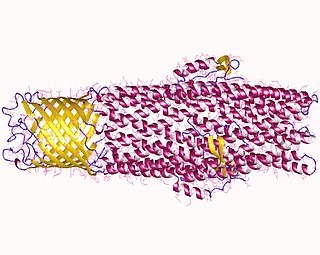
All microorganisms, with a few exceptions, have highly conserved DNA sequences in their genome that are transcribed and translated to efflux pumps. Efflux pumps are capable of moving a variety of different toxic compounds out of cells, such as antibiotics, heavy metals, organic pollutants, plant-produced compounds, quorum sensing signals, bacterial metabolites and neurotransmitters via active efflux, which is vital part for xenobiotic metabolism. This active efflux mechanism is responsible for various types of resistance to bacterial pathogens within bacterial species - the most concerning being antibiotic resistance because microorganisms can have adapted efflux pumps to divert toxins out of the cytoplasm and into extracellular media.

Acinetobacter baumannii is a typically short, almost round, rod-shaped (coccobacillus) Gram-negative bacterium. It is named after the bacteriologist Paul Baumann. It can be an opportunistic pathogen in humans, affecting people with compromised immune systems, and is becoming increasingly important as a hospital-derived (nosocomial) infection. While other species of the genus Acinetobacter are often found in soil samples, it is almost exclusively isolated from hospital environments. Although occasionally it has been found in environmental soil and water samples, its natural habitat is still not known.
Cross-resistance is when something develops resistance to several substances that have a similar mechanism of action. For example, say a certain type of bacteria develops resistance to one antibiotic. That bacteria then also has resistance to several other antibiotics that target the same protein or use the same route to get into the bacterium. A real example of cross-resistance occurred for nalidixic acid and ciprofloxacin, which are both quinolone antibiotics. When bacteria developed resistance to ciprofloxacin, they also developed resistance to nalidixic acid because both drugs work by inhibiting topoisomerase, a key enzyme in DNA replication. Due to cross-resistance, antimicrobial treatments like phage therapy can quickly lose their efficacy against bacteria.
The resistome has been used to describe to two similar yet separate concepts:

Functional cloning is a molecular cloning technique that relies on prior knowledge of the encoded protein’s sequence or function for gene identification. In this assay, a genomic or cDNA library is screened to identify the genetic sequence of a protein of interest. Expression cDNA libraries may be screened with antibodies specific for the protein of interest or may rely on selection via the protein function. Historically, the amino acid sequence of a protein was used to prepare degenerate oligonucleotides which were then probed against the library to identify the gene encoding the protein of interest. Once candidate clones carrying the gene of interest are identified, they are sequenced and their identity is confirmed. This method of cloning allows researchers to screen entire genomes without prior knowledge of the location of the gene or the genetic sequence.

Microbiota are the range of microorganisms that may be commensal, symbiotic, or pathogenic found in and on all multicellular organisms, including plants. Microbiota include bacteria, archaea, protists, fungi, and viruses, and have been found to be crucial for immunologic, hormonal, and metabolic homeostasis of their host.

Antibiotic use in livestock is the use of antibiotics for any purpose in the husbandry of livestock, which includes treatment when ill (therapeutic), treatment of a group of animals when at least one is diagnosed with clinical infection (metaphylaxis), and preventative treatment (prophylaxis). Antibiotics are an important tool to treat animal as well as human disease, safeguard animal health and welfare, and support food safety. However, used irresponsibly, this may lead to antibiotic resistance which may impact human, animal and environmental health.
Mustard is a database that tracks Antimicrobial Resistance Determinants (ARDs). The method by which it tracks ARDs is using their own method adapted from Protein Homology Modelling called Pairwise Comparative Modelling (PCM), which increase specificity protein prediction, especially for distantly related protein homologues. Using PCM, 6095 ARDs from 20 families in the human gut microbiota. Antibiotic resistance databases used were ResFinder, ARG-ANNOT, the now defunct Lahey Clinic, Marilyn Roberts website for tetracycline and macrolide resistance genes and metagenomics.
Clinical metagenomic next-generation sequencing (mNGS) is the comprehensive analysis of microbial and host genetic material in clinical samples from patients. It uses the techniques of metagenomics to identify and characterize the genome of bacteria, fungi, parasites, and viruses without the need for a prior knowledge of a specific pathogen directly from clinical specimens. The capacity to detect all the potential pathogens in a sample makes metagenomic next generation sequencing a potent tool in the diagnosis of infectious disease especially when other more directed assays, such as PCR, fail. Its limitations include clinical utility, laboratory validity, sense and sensitivity, cost and regulatory considerations.

Multidrug-resistant bacteria are bacteria that are resistant to three or more classes of antimicrobial drugs. MDR bacteria have seen an increase in prevalence in recent years and pose serious risks to public health. MDR bacteria can be broken into 3 main categories: Gram-positive, Gram-negative, and other (acid-stain). These bacteria employ various adaptations to avoid or mitigate the damage done by antimicrobials. With increased access to modern medicine there has been a sharp increase in the amount of antibiotics consumed. Given the abundant use of antibiotics there has been a considerable increase in the evolution of antimicrobial resistance factors, now outpacing the development of new antibiotics.