
Pseudomonas is a genus of Gram-negative, Gammaproteobacteria, belonging to the family Pseudomonadaceae and containing 191 validly described species. The members of the genus demonstrate a great deal of metabolic diversity and consequently are able to colonize a wide range of niches. Their ease of culture in vitro and availability of an increasing number of Pseudomonas strain genome sequences has made the genus an excellent focus for scientific research; the best studied species include P. aeruginosa in its role as an opportunistic human pathogen, the plant pathogen P. syringae, the soil bacterium P. putida, and the plant growth-promoting P. fluorescens, P. lini, P. migulae, and P. graminis.
Pseudomonas putida is a Gram-negative, rod-shaped, saprotrophic soil bacterium. Based on 16S rRNA analysis, P. putida was taxonomically confirmed to be a Pseudomonas species and placed, along with several other species, in the P. putida group, to which it lends its name.
Acyl-CoA dehydrogenases (ACADs) are a class of enzymes that function to catalyze the initial step in each cycle of fatty acid β-oxidation in the mitochondria of cells. Their action results in the introduction of a trans double-bond between C2 (α) and C3 (β) of the acyl-CoA thioester substrate Flavin adenine dinucleotide (FAD) is a required co-factor in addition to the presence of an active site glutamate in order for the enzyme to function.

4-hydroxyphenylacetate 3-monooxygenase (EC 1.14.14.9) is an enzyme that catalyzes the chemical reaction
In enzymology, an acetate CoA-transferase is an enzyme that catalyzes the chemical reaction
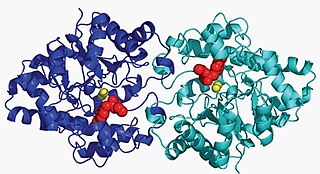
Aryldialkylphosphatase is a metalloenzyme that hydrolyzes the triester linkage found in organophosphate insecticides.

Pyocyanin (PCN−) is one of the many toxins produced and secreted by the Gram negative bacterium Pseudomonas aeruginosa. Pyocyanin is a blue, secondary metabolite with the ability to oxidise and reduce other molecules and therefore kill microbes competing against P. aeruginosa as well as mammalian cells of the lungs which P. aeruginosa has infected during cystic fibrosis. Since pyocyanin is a zwitterion at blood pH, it is easily able to cross the cell membrane. There are three different states in which pyocyanin can exist: oxidized, monovalently reduced or divalently reduced. Mitochondria play an important role in the cycling of pyocyanin between its redox states. Due to its redox-active properties, pyocyanin generates reactive oxygen species.

Cobalamin biosynthesis is the process by which bacteria and archea make cobalamin, vitamin B12. Many steps are involved in converting aminolevulinic acid via uroporphyrinogen III and adenosylcobyric acid to the final forms in which it is used by enzymes in both the producing organisms and other species, including humans who acquire it through their diet.

Rhamnolipids are a class of glycolipid produced by Pseudomonas aeruginosa, amongst other organisms, frequently cited as the best characterised of the bacterial surfactants. They have a glycosyl head group, in this case a rhamnose moiety, and a 3-(hydroxyalkanoyloxy)alkanoic acid (HAA) fatty acid tail, such as 3-hydroxydecanoic acid.
The Nucleobase cation symporter-2 (NCS2) family, also called the Nucleobase ascorbate transporter (NAT) family, consists of over 1000 sequenced proteins derived from gram-negative and gram-positive bacteria, archaea, fungi, plants and animals. The NCS2/NAT family is a member of the APC Superfamily of secondary carriers. Of the five known families of transporters that act on nucleobases, NCS2/NAT is the only one that is most widespread. Many functionally characterized members are specific for nucleobases including both purines and pyrimidines, but others are purine-specific. However, two closely related rat/human members of the family, SVCT1 and SVCT2, localized to different tissues of the body, co-transport L-ascorbate (vitamin C) and Na+ with a high degree of specificity and high affinity for the vitamin. Clustering of NCS2/NAT family members on the phylogenetic tree is complex, with bacterial proteins and eukaryotic proteins each falling into at least three distinct clusters. The plant and animal proteins cluster loosely together, but the fungal proteins branch from one of the three bacterial clusters forming a tighter grouping. E. coli possesses four distantly related paralogous members of the NCS2 family.
Acetoin dehydrogenase (EC 2.3.1.190, acetoin dehydrogenase complex, acetoin dehydrogenase enzyme system, AoDH ES) is an enzyme with systematic name acetyl-CoA:acetoin O-acetyltransferase. This enzyme catalyses the following chemical reaction
2-Hydroxymuconate-6-semialdehyde dehydrogenase (EC 1.2.1.85, xylG [gene], praB [gene] ) is an enzyme with systematic name (2E,4Z)-2-hydroxy-6-oxohexa-2,4-dienoate:NAD+ oxidoreductase. This enzyme catalyses the following chemical reaction
(2,2,3-Trimethyl-5-oxocyclopent-3-enyl)acetyl-CoA 1,5-monooxygenase (EC 1.14.13.160, 2-oxo-Delta3-4,5,5-trimethylcyclopentenylacetyl-CoA monooxygenase, 2-oxo-Delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase, OTEMO) is an enzyme with systematic name ((1R)-2,2,3-trimethyl-5-oxocyclopent-3-enyl)acetyl-CoA,NADPH:oxygen oxidoreductase (1,5-lactonizing). This enzyme catalyses the following chemical reaction
Beta-ketodecanoyl-(acyl-carrier-protein) synthase (EC 2.3.1.207) is an enzyme with systematic name octanoyl-CoA:malonyl-(acyl-carrier protein) C-heptanoylltransferase (decarboxylating, CoA-forming). This enzyme catalyses the following chemical reaction
3-Fumarylpyruvate hydrolase (EC 3.7.1.20, nagK (gene), naaD (gene)) is an enzyme with systematic name 3-fumarylpyruvate hydrolyase. This enzyme catalyses the following chemical reaction
2-hydroxychromene-2-carboxylate isomerase is an enzyme with systematic name 2-hydroxy-2H-chromene-2-carboxylate---(3E)-4-(2-hydroxyphenyl)-2-oxobut-3-enoate isomerase. This enzyme catalyses the following chemical reaction
Thauera aromatica is a species of bacteria. Its type strain is K 172T.
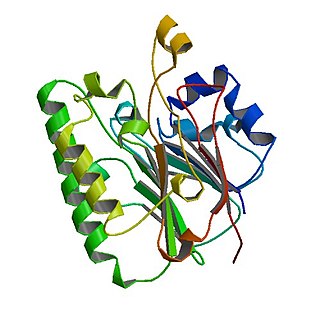
The Catabolite repression control (Crc) protein participates in suppressing expression of several genes involved in utilization of carbon sources in Pseudomonas bacteria. Presence of organic acids triggers activation of Crc and in conjunction with the Hfq protein genes that metabolize a given carbon source are downregulated until another more favorable carbon source is depleted. Crc-mediated regulation impact processes such as biofilm formation, virulence and antibiotic susceptibility.
The 4-Toluene Sulfonate Uptake Permease (TSUP) family is also referred to as the TauE/SafE/YfcA/DUF81 Family. Although its members have not been rigorously characterized, evidence is available that at least some members function in the transport of sulfur containing organic compounds. These include 4-toluene sulfonate which may be transported by the TsaS of Cupriavidus necator, sulfolactate which may be exported by the TauE protein of Cupriavidus necator and sulfoacetate which may be exported by the SafE1 protein of Neptuniibacter caesariensis. Another member of the TSUP family, TsaS of Comamonas testosteroni, has been reported to function in the uptake of 4-toluene sulfonate. None of these functional assignments can be considered to be certain.

The Phosphate (Pho) regulon is a bacterial regulatory mechanism used for the conservation and management of inorganic phosphate within the cell. It was first discovered in Escherichia coli as an operating system for the bacterial strain, and was later identified in other species. The Pho system is composed of various components including extracellular enzymes and transporters that are capable of phosphate assimilation in addition to extracting inorganic phosphate from organic sources. This is an essential process since phosphate plays an important role in cellular membranes, genetic expression, and metabolism within the cell. Under low nutrient availability, the Pho regulon helps the cell survive and thrive despite a depletion of phosphate within the environment. When this occurs, phosphate starvation-inducible (psi) genes activate other proteins that aid in the transport of inorganic phosphate.








