
A bacteriophage, also known informally as a phage, is a virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν, meaning "to devour". Bacteriophages are composed of proteins that encapsulate a DNA or RNA genome, and may have structures that are either simple or elaborate. Their genomes may encode as few as four genes and as many as hundreds of genes. Phages replicate within the bacterium following the injection of their genome into its cytoplasm.

A prophage is a bacteriophage genome that is integrated into the circular bacterial chromosome or exists as an extrachromosomal plasmid within the bacterial cell. Integration of prophages into the bacterial host is the characteristic step of the lysogenic cycle of temperate phages. Prophages remain latent in the genome through multiple cell divisions until activation by an external factor, such as UV light, leading to production of new phage particles that will lyse the cell and spread. As ubiquitous mobile genetic elements, prophages play important roles in bacterial genetics and evolution, such as in the acquisition of virulence factors.

Metagenomics is the study of genetic material recovered directly from environmental or clinical samples by a method called sequencing. The broad field may also be referred to as environmental genomics, ecogenomics, community genomics or microbiomics.

Filamentous bacteriophages are a family of viruses (Inoviridae) that infect bacteria, or bacteriophages. They are named for their filamentous shape, a worm-like chain, about 6 nm in diameter and about 1000-2000 nm long. This distinctive shape reflects their method of replication: the coat of the virion comprises five types of viral protein, which are located in the inner membrane of the host bacterium during phage assembly, and these proteins are added to the nascent virion's DNA as it is extruded through the membrane. The simplicity of filamentous phages makes them an appealing model organism for research in molecular biology, and they have also shown promise as tools in nanotechnology and immunology.

Marine microorganisms are defined by their habitat as microorganisms living in a marine environment, that is, in the saltwater of a sea or ocean or the brackish water of a coastal estuary. A microorganism is any microscopic living organism or virus, which is invisibly small to the unaided human eye without magnification. Microorganisms are very diverse. They can be single-celled or multicellular and include bacteria, archaea, viruses, and most protozoa, as well as some fungi, algae, and animals, such as rotifers and copepods. Many macroscopic animals and plants have microscopic juvenile stages. Some microbiologists also classify viruses as microorganisms, but others consider these as non-living.
The hologenome theory of evolution recasts the individual animal or plant as a community or a "holobiont" – the host plus all of its symbiotic microbes. Consequently, the collective genomes of the holobiont form a "hologenome". Holobionts and hologenomes are structural entities that replace misnomers in the context of host-microbiota symbioses such as superorganism, organ, and metagenome. Variation in the hologenome may encode phenotypic plasticity of the holobiont and can be subject to evolutionary changes caused by selection and drift, if portions of the hologenome are transmitted between generations with reasonable fidelity. One of the important outcomes of recasting the individual as a holobiont subject to evolutionary forces is that genetic variation in the hologenome can be brought about by changes in the host genome and also by changes in the microbiome, including new acquisitions of microbes, horizontal gene transfers, and changes in microbial abundance within hosts. Although there is a rich literature on binary host–microbe symbioses, the hologenome concept distinguishes itself by including the vast symbiotic complexity inherent in many multicellular hosts.
Biological dark matter is an informal term for unclassified or poorly understood genetic material. This genetic material may refer to genetic material produced by unclassified microorganisms. By extension, biological dark matter may also refer to the un-isolated microorganisms whose existence can only be inferred from the genetic material that they produce. Some of the genetic material may not fall under the three existing domains of life: Bacteria, Archaea and Eukaryota; thus, it has been suggested that a possible fourth domain of life may yet be discovered, although other explanations are also probable. Alternatively, the genetic material may refer to non-coding DNA and non-coding RNA produced by known organisms.

The human virome is the total collection of viruses in and on the human body. Viruses in the human body may infect both human cells and other microbes such as bacteria. Some viruses cause disease, while others may be asymptomatic. Certain viruses are also integrated into the human genome as proviruses or endogenous viral elements.

Viral metagenomics uses metagenomic technologies to detect viral genomic material from diverse environmental and clinical samples. Viruses are the most abundant biological entity and are extremely diverse; however, only a small fraction of viruses have been sequenced and only an even smaller fraction have been isolated and cultured. Sequencing viruses can be challenging because viruses lack a universally conserved marker gene so gene-based approaches are limited. Metagenomics can be used to study and analyze unculturable viruses and has been an important tool in understanding viral diversity and abundance and in the discovery of novel viruses. For example, metagenomics methods have been used to describe viruses associated with cancerous tumors and in terrestrial ecosystems.

CrAss-like phage (crassviruses) are a bacteriophage family representing the most abundant viruses in the human gut, discovered in 2014 by cross assembling reads in human fecal metagenomes. In silico comparative genomics and taxonomic analysis have found that crAss-like phages represent a highly abundant and diverse family of viruses. CrAss-like phage were predicted to infect bacteria of the Bacteroidota phylum and the prediction was later confirmed when the first crAss-like phage (crAss001) was isolated on a Bacteroidota host in 2018. Crassviruses are podoviruses, possessing short non-contractile tails and icosahedral capsids. The first 3D structure of a crassvirus was determined by cryo-EM in 2023. While the presence of crAss-like phage in the human gut is not yet associated with any specific health condition, they are generally associated with a healthy gut microbiome and likely impact significantly on the gut Bacteroidota.

A microbiome is the community of microorganisms that can usually be found living together in any given habitat. It was defined more precisely in 1988 by Whipps et al. as "a characteristic microbial community occupying a reasonably well-defined habitat which has distinct physio-chemical properties. The term thus not only refers to the microorganisms involved but also encompasses their theatre of activity". In 2020, an international panel of experts published the outcome of their discussions on the definition of the microbiome. They proposed a definition of the microbiome based on a revival of the "compact, clear, and comprehensive description of the term" as originally provided by Whipps et al., but supplemented with two explanatory paragraphs, the first pronouncing the dynamic character of the microbiome, and the second clearly separating the term microbiota from the term microbiome.

A holobiont is an assemblage of a host and the many other species living in or around it, which together form a discrete ecological unit through symbiosis, though there is controversy over this discreteness. The components of a holobiont are individual species or bionts, while the combined genome of all bionts is the hologenome. The holobiont concept was initially introduced by the German theoretical biologist Adolf Meyer-Abich in 1943, and then apparently independently by Dr. Lynn Margulis in her 1991 book Symbiosis as a Source of Evolutionary Innovation. The concept has evolved since the original formulations. Holobionts include the host, virome, microbiome, and any other organisms which contribute in some way to the functioning of the whole. Well-studied holobionts include reef-building corals and humans.

Virome refers to the assemblage of viruses that is often investigated and described by metagenomic sequencing of viral nucleic acids that are found associated with a particular ecosystem, organism or holobiont. The word is frequently used to describe environmental viral shotgun metagenomes. Viruses, including bacteriophages, are found in all environments, and studies of the virome have provided insights into nutrient cycling, development of immunity, and a major source of genes through lysogenic conversion. Also, the human virome has been characterized in nine organs of 31 Finnish individuals using qPCR and NGS methodologies.

The "Kill the Winner" hypothesis (KtW) is an ecological model of population growth involving prokaryotes, viruses and protozoans that links trophic interactions to biogeochemistry. The model is related to the Lotka–Volterra equations. It assumes that prokaryotes adopt one of two strategies when competing for limited resources: priority is either given to population growth ("winners") or survival ("defenders"). As "winners" become more abundant and active in their environment, their contact with host-specific viruses increases, making them more susceptible to viral infection and lysis. Thus, viruses moderate the population size of "winners" and allow multiple species to coexist. Current understanding of KtW primarily stems from studies of lytic viruses and their host populations.
Auxiliary metabolic genes (AMGs) are found in many bacteriophages but originated in bacterial cells. AMGs modulate host cell metabolism during infection so that the phage can replicate more efficiently. For instance, bacteriophages that infect the abundant marine cyanobacteria Synechococcus and Prochlorococcus (cyanophages) carry AMGs that have been acquired from their immediate host as well as more distantly-related bacteria. Cyanophage AMGs support a variety of functions including photosynthesis, carbon metabolism, nucleic acid synthesis and metabolism. AMGs also have broader ecological impacts beyond their host including their influence on biogeochemical cycling.

Mya Breitbart is an American biologist and professor of biological oceanography at the University of South Florida's College of Marine Science. She is best known for her contributions to the field of viral metagenomics. Popular Science recognized her because of her approach of not trying to sequence individual viruses or organisms but to sequence everything in a given ecosystem.

All animals on Earth form associations with microorganisms, including protists, bacteria, archaea, fungi, and viruses. In the ocean, animal–microbial relationships were historically explored in single host–symbiont systems. However, new explorations into the diversity of marine microorganisms associating with diverse marine animal hosts is moving the field into studies that address interactions between the animal host and a more multi-member microbiome. The potential for microbiomes to influence the health, physiology, behavior, and ecology of marine animals could alter current understandings of how marine animals adapt to change, and especially the growing climate-related and anthropogenic-induced changes already impacting the ocean environment.
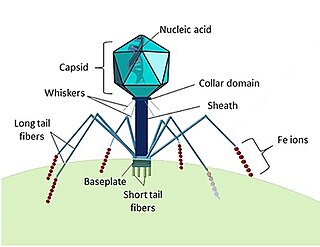
Marine viruses are defined by their habitat as viruses that are found in marine environments, that is, in the saltwater of seas or oceans or the brackish water of coastal estuaries. Viruses are small infectious agents that can only replicate inside the living cells of a host organism, because they need the replication machinery of the host to do so. They can infect all types of life forms, from animals and plants to microorganisms, including bacteria and archaea.
Diversity-generating retroelements (DGRs) are a family of retroelements that were first found in Bordetella phage (BPP-1), and since been found in bacteria, Archaea, Archaean viruses, temperate phages, and lytic phages. DGRs benefit their host by mutating particular regions of specific target proteins, for instance, phage tail fiber in BPP-1, lipoprotein in legionella pneumophila, and TvpA in Treponema denticola . An error-prone reverse transcriptase is responsible for generating these hypervariable regions in target proteins. In mutagenic retrohoming, a mutagenized cDNA is reverse transcribed from a template region (TR), and is replaced with a segment similar to the template region called variable region (VR). Accessory variability determinant (Avd) protein is another component of DGRs, and its complex formation with the error-prone RT is of importance to mutagenic rehoming.

A phageome is a community of bacteriophages and their metagenomes localized in a particular environment, similar to a microbiome. The term was first used in an article by Modi et al in 2013 and has continued to be used in scientific articles that relate to bacteriophages and their metagenomes. A bacteriophage, or phage for short, is a virus that has the ability to infect bacteria and archaea, and can replicate inside of them. Phageome is a subcategory of virome, which is all of the viruses that are associated with a host or environment. Phages make up the majority of most viromes and are currently understood as being the most abundant organism. Oftentimes scientists will look only at a phageome instead of a virome while conducting research.













