
In molecular biology, a transcription factor (TF) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the right cell at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome.

A homeobox is a DNA sequence, around 180 base pairs long, found within genes that are involved in the regulation of patterns of anatomical development (morphogenesis) in animals, fungi, plants, and numerous single cell eukaryotes. Homeobox genes encode homeodomain protein products that are transcription factors sharing a characteristic protein fold structure that binds DNA to regulate expression of target genes. Homeodomain proteins regulate gene expression and cell differentiation during early embryonic development, thus mutations in homeobox genes can cause developmental disorders.

A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions (Zn2+) in order to stabilize the fold. It was originally coined to describe the finger-like appearance of a hypothesized structure from the African clawed frog (Xenopus laevis) transcription factor IIIA. However, it has been found to encompass a wide variety of differing protein structures in eukaryotic cells. Xenopus laevis TFIIIA was originally demonstrated to contain zinc and require the metal for function in 1983, the first such reported zinc requirement for a gene regulatory protein followed soon thereafter by the Krüppel factor in Drosophila. It often appears as a metal-binding domain in multi-domain proteins.
FOXproteins are a family of transcription factors that play important roles in regulating the expression of genes involved in cell growth, proliferation, differentiation, and longevity. Many FOX proteins are important to embryonic development. FOX proteins also have pioneering transcription activity by being able to bind condensed chromatin during cell differentiation processes.
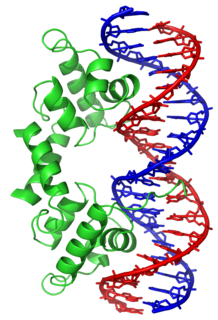
In proteins, the helix-turn-helix (HTH) is a major structural motif capable of binding DNA. Each monomer incorporates two α helices, joined by a short strand of amino acids, that bind to the major groove of DNA. The HTH motif occurs in many proteins that regulate gene expression. It should not be confused with the helix–loop–helix motif.

A morphogen is a substance whose non-uniform distribution governs the pattern of tissue development in the process of morphogenesis or pattern formation, one of the core processes of developmental biology, establishing positions of the various specialized cell types within a tissue. More specifically, a morphogen is a signaling molecule that acts directly on cells to produce specific cellular responses depending on its local concentration.

A basic helix–loop–helix (bHLH) is a protein structural motif that characterizes one of the largest families of dimerizing transcription factors.
Hox genes, a subset of homeobox genes, are a group of related genes that specify regions of the body plan of an embryo along the head-tail axis of animals. Hox proteins encode and specify the characteristics of 'position', ensuring that the correct structures form in the correct places of the body. For example, Hox genes in insects specify which appendages form on a segment, and Hox genes in vertebrates specify the types and shape of vertebrae that will form. In segmented animals, Hox proteins thus confer segmental or positional identity, but do not form the actual segments themselves.
An E-box is a DNA response element found in some eukaryotes that acts as a protein-binding site and has been found to regulate gene expression in neurons, muscles, and other tissues. Its specific DNA sequence, CANNTG, with a palindromic canonical sequence of CACGTG, is recognized and bound by transcription factors to initiate gene transcription. Once the transcription factors bind to the promoters through the E-box, other enzymes can bind to the promoter and facilitate transcription from DNA to mRNA.

In molecular biology, heat shock factors (HSF), are the transcription factors that regulate the expression of the heat shock proteins. A typical example is the heat shock factor of Drosophila melanogaster.

Protein C-ets-1 is a protein that in humans is encoded by the ETS1 gene. The protein encoded by this gene belongs to the ETS family of transcription factors.

DNA-binding protein inhibitor ID-2 is a protein that in humans is encoded by the ID2 gene.

Transcription factor HES1 is a protein that is encoded by the Hes1 gene, and is the mammalian homolog of the hairy gene in Drosophila. HES1 is one of the seven members of the Hes gene family (HES1-7). Hes genes code nuclear proteins that suppress transcription.

Transcription factor AP-4 , also known as TFAP4, is a protein which in humans is encoded by the TFAP4 gene.

Forkhead box protein A2 (FOXA2), also known as hepatocyte nuclear factor 3-beta (HNF-3B), is a transcription factor that plays an important role during development, in mature tissues and, when dysregulated or mutated, also in cancer.

Hepatocyte nuclear factor 3-gamma (HNF-3G), also known as forkhead box protein A3 (FOXA3) or transcription factor 3G (TCF-3G) is a protein that in humans is encoded by the FOXA3 gene.

In molecular biology the DM domain is a protein domain first discovered in the doublesex proteins of Drosophila melanogaster and is also seen in C. elegans and mammalian proteins. In D. melanogaster the doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. These proteins are believed to function as transcription factors on downstream sex-determination genes, especially on neuroblast differentiation and yolk protein genes transcription.
In molecular biology, the BESS domain is a protein domain which has been named after the three proteins that originally defined the domain: BEAF, Suvar(3)7 and Stonewall ). The BESS domain is 40 amino acid residues long and is predicted to be composed of three alpha helices, as such it might be related to the myb/SANT HTH domain. The BESS domain directs a variety of protein-protein interactions, including interactions with itself, with Dorsal, and with a TBP-associated factor. It is found in a single copy in Drosophila proteins and is often associated with the MADF domain.
Pioneer factors are transcription factors that can directly bind condensed chromatin. They can have positive and negative effects on transcription and are important in recruiting other transcription factors and histone modification enzymes as well as controlling DNA methylation. They were first discovered in 2002 as factors capable of binding to target sites on nucleosomal DNA in compacted chromatin and endowing competency for gene activity during hepatogenesis. Pioneer factors are involved in initiating cell differentiation and activation of cell-specific genes. This property is observed in histone fold-domain containing transcription factors and other transcription factors that use zinc finger(s) for DNA binding.

Forkhead box D1 is a protein that in humans is encoded by the FOXD1 gene. Forkhead d1 is a kidney expressed transcription factor maps at the chromosome 5 at position 5q12—q13, identified in Drosophila forkhead protein and mammalian HNF3 transcription factor. The name of was derived from two spiked head structures in the embryos of Drosophila forkhead mutant. It belong to transcription factor family that displays remarkable functional diversity and involved in a wide variety of biological processes. The most commonly used synonyms for Forkhead D1 are, FOX D1, FREAC-4 and BF2.















