Related Research Articles

The human genome is a complete set of nucleic acid sequences for humans, encoded as DNA within the 23 chromosome pairs in cell nuclei and in a small DNA molecule found within individual mitochondria. These are usually treated separately as the nuclear genome and the mitochondrial genome. Human genomes include both protein-coding DNA genes and noncoding DNA. Haploid human genomes, which are contained in germ cells consist of three billion DNA base pairs, while diploid genomes have twice the DNA content. While there are significant differences among the genomes of human individuals, these are considerably smaller than the differences between humans and their closest living relatives, the bonobos and chimpanzees.
Non-coding DNA sequences are components of an organism's DNA that do not encode protein sequences. Some non-coding DNA is transcribed into functional non-coding RNA molecules. Other functions of non-coding DNA include the transcriptional and translational regulation of protein-coding sequences, scaffold attachment regions, origins of DNA replication, centromeres and telomeres. Its RNA counterpart is non-coding RNA.

A Barr body or X-chromatin is an inactive X chromosome in a cell with more than one X chromosome, rendered inactive in a process called lyonization, in species with XY sex-determination. The Lyon hypothesis states that in cells with multiple X chromosomes, all but one are inactivated during mammalian embryogenesis. This happens early in embryonic development at random in mammals, except in marsupials and in some extra-embryonic tissues of some placental mammals, in which the X chromosome from the sperm is always deactivated.
Heterochromatin is a tightly packed form of DNA or condensed DNA, which comes in multiple varieties. These varieties lie on a continuum between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a role in the expression of genes. Because it is tightly packed, it was thought to be inaccessible to polymerases and therefore not transcribed; however, according to Volpe et al. (2002), and many other papers since, much of this DNA is in fact transcribed, but it is continuously turned over via RNA-induced transcriptional silencing (RITS). Recent studies with electron microscopy and OsO4 staining reveal that the dense packing is not due to the chromatin.
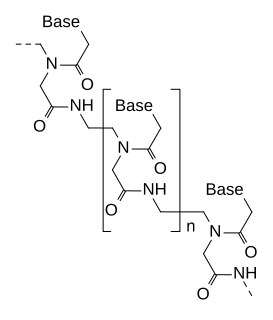
Peptide nucleic acid (PNA) is an artificially synthesized polymer similar to DNA or RNA.

A non-coding RNA (ncRNA) is an RNA molecule that is not translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, exRNAs, scaRNAs and the long ncRNAs such as Xist and HOTAIR.

Dosage compensation is the process by which organisms equalize the expression of genes between members of different biological sexes. Across species, different sexes are often characterized by different types and numbers of sex chromosomes. In order to neutralize the large difference in gene dosage produced by differing numbers of sex chromosomes among the sexes, various evolutionary branches have acquired various methods to equalize gene expression among the sexes. Because sex chromosomes contain different numbers of genes, different species of organisms have developed different mechanisms to cope with this inequality. Replicating the actual gene is impossible; thus organisms instead equalize the expression from each gene. For example, in humans, females (XX) silence the transcription of one X chromosome of each pair, and transcribe all information from the other, expressed X chromosome. Thus, human females have the same number of expressed X-linked genes as do human males (XY), both sexes having essentially one X chromosome per cell, from which to transcribe and express genes.

X-inactivation is a process by which one of the copies of the X chromosome is inactivated in therian female mammals. The inactive X chromosome is silenced by it being packaged into a transcriptionally inactive structure called heterochromatin. As nearly all female mammals have two X chromosomes, X-inactivation prevents them from having twice as many X chromosome gene products as males, who only possess a single copy of the X chromosome.

Potassium voltage-gated channel subfamily A member 1 also known as Kv1.1 is a shaker related voltage-gated potassium channel that in humans is encoded by the KCNA1 gene. The Isaacs syndrome is a result of an autoimmune reaction against the Kv1.1 ion channel.

Xist is a non-coding RNA on the X chromosome of the placental mammals that acts as a major effector of the X-inactivation process. It is a component of the Xic – X-chromosome inactivation centre – along with two other RNA genes and two protein genes.

Long non-coding RNAs are a type of RNA, generally defined as transcripts more than 200 nucleotides that are not translated into protein. This arbitrary limit distinguishes long ncRNAs from small non-coding RNAs, such as microRNAs (miRNAs), small interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), and other short RNAs. Long intervening/intergenic noncoding RNAs (lincRNAs) are sequences of lncRNA which do not overlap protein-coding genes.

HOTAIR is a human gene located between HOXC11 and HOXC12 on chromosome 12. It is the first example of an RNA expressed on one chromosome that has been found to influence transcription of HOXD cluster posterior genes located on chromosome 2. The sequence and function of HOTAIR is different in human and mouse. Sequence analysis of HOTAIR revealed that it exists in mammals, has poorly conserved sequences and considerably conserved structures, and has evolved faster than nearby HoxC genes. A subsequent study identified HOTAIR has 32 nucleotide long conserved noncoding element (CNE) that has a paralogous copy in HOXD cluster region, suggesting that the HOTAIR conserved sequences predates whole genome duplication events at the root of vertebrate. While the conserved sequence paralogous with HOXD cluster is 32 nucleotide long, the HOTAIR sequence conserved from human to fish is about 200 nucleotide long and is marked by active enhancer features.
Skewed X-chromosome inactivation occurs when the X-inactivation of one X chromosome is favored over the other, leading to an uneven number of cells with each chromosome inactivated. It is usually defined as one allele being found on the active X chromosome in over 75% of cells, and extreme skewing is when over 90% of cells have inactivated the same X chromosome. It can be caused by primary nonrandom inactivation, either by chance due to a small cell pool or directed by genes, or by secondary nonrandom inactivation, which occurs by selection.

KCNQ1 downstream neighbour (KCNQ1DN) is a long non-coding RNA gene. In humans, it is located on chromosome 11p15.5 between the CDKN1C and KCNQ1 genes. It is an imprinted gene, expressed from the maternal allele. Reduced expression of KCNQ1DN is observed in Wilms' tumours.

In molecular biology, JPX transcript, XIST activator, also known as Jpx, is a long non-coding RNA. In humans, it is located on the X chromosome. It was identified during sequence analysis of the X inactivation centre, surrounding the Xist gene. Jpx upregulates expression of Xist.

Tsix is a non-coding RNA gene that is antisense to the Xist RNA. Tsix binds Xist during X chromosome inactivation. The name Tsix comes from the reverse of Xist, which stands for X-inactive specific transcript.
Development before birth, including gametogenesis, embryogenesis, and fetal development, is the process of body development from the gametes are formed to eventually combine into a zygote to when the fully developed organism exits the uterus. Epigenetic processes are vital to fetal development due to the need to differentiate from a single cell to a variety of cell types that are arranged in such a way to produce cohesive tissues, organs, and systems.

X chromosome inactivation (XCI) is the phenomenon that has been selected during the evolution to balance X-linked gene dosage between XX females and XY males.
Carolyn J. Brown is a Canadian geneticist and Professor in the Department of Medical Genetics at the University of British Columbia. Brown is known for her studies on X-chromosome inactivation, having discovered the human XIST gene in 1990.

Chromosome X Open Reading Frame 38 (CXorf38) is a protein which, in humans, is encoded by the CXorf38 gene. CXorf38 appears in multiple studies regarding the escape of X chromosome inactivation.
References
- ↑ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ↑ Chureau C, Prissette M, Bourdet A, Barbe V, Cattolico L, Jones L, et al. (2002). "Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine". Genome Res. 12 (6): 894–908. doi:10.1101/gr.152902. PMC 1383731 . PMID 12045143.
- 1 2 Chureau C, Chantalat S, Romito A, Galvani A, Duret L, Avner P, et al. (2011). "Ftx is a non-coding RNA which affects Xist expression and chromatin structure within the X-inactivation center region". Hum Mol Genet. 20 (4): 705–718. doi: 10.1093/hmg/ddq516 . PMID 21118898.