Related Research Articles

In molecular biology, a transcription factor (TF) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome.

A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions (Zn2+) in order to stabilize the fold. It was originally coined to describe the finger-like appearance of a hypothesized structure from the African clawed frog (Xenopus laevis) transcription factor IIIA. However, it has been found to encompass a wide variety of differing protein structures in eukaryotic cells. Xenopus laevis TFIIIA was originally demonstrated to contain zinc and require the metal for function in 1983, the first such reported zinc requirement for a gene regulatory protein followed soon thereafter by the Krüppel factor in Drosophila. It often appears as a metal-binding domain in multi-domain proteins.
In molecular biology, the TATA box is a sequence of DNA found in the core promoter region of genes in archaea and eukaryotes. The bacterial homolog of the TATA box is called the Pribnow box which has a shorter consensus sequence.
A transcriptional activator is a protein that increases transcription of a gene or set of genes. Activators are considered to have positive control over gene expression, as they function to promote gene transcription and, in some cases, are required for the transcription of genes to occur. Most activators are DNA-binding proteins that bind to enhancers or promoter-proximal elements. The DNA site bound by the activator is referred to as an "activator-binding site". The part of the activator that makes protein–protein interactions with the general transcription machinery is referred to as an "activating region" or "activation domain".

Histone acetyltransferases (HATs) are enzymes that acetylate conserved lysine amino acids on histone proteins by transferring an acetyl group from acetyl-CoA to form ε-N-acetyllysine. DNA is wrapped around histones, and, by transferring an acetyl group to the histones, genes can be turned on and off. In general, histone acetylation increases gene expression.
In molecular biology, a CCAAT box is a distinct pattern of nucleotides with GGCCAATCT consensus sequence that occur upstream by 60–100 bases to the initial transcription site. The CAAT box signals the binding site for the RNA transcription factor, and is typically accompanied by a conserved consensus sequence. It is an invariant DNA sequence at about minus 70 base pairs from the origin of transcription in many eukaryotic promoters. Genes that have this element seem to require it for the gene to be transcribed in sufficient quantities. It is frequently absent from genes that encode proteins used in virtually all cells. This box along with the GC box is known for binding general transcription factors. Both of these consensus sequences belong to the regulatory promoter. Full gene expression occurs when transcription activator proteins bind to each module within the regulatory promoter. Protein specific binding is required for the CCAAT box activation. These proteins are known as CCAAT box binding proteins/CCAAT box binding factors.

Catabolite activator protein is a trans-acting transcriptional activator that exists as a homodimer in solution. Each subunit of CAP is composed of a ligand-binding domain at the N-terminus and a DNA-binding domain at the C-terminus. Two cAMP molecules bind dimeric CAP with negative cooperativity. Cyclic AMP functions as an allosteric effector by increasing CAP's affinity for DNA. CAP binds a DNA region upstream from the DNA binding site of RNA Polymerase. CAP activates transcription through protein-protein interactions with the α-subunit of RNA Polymerase. This protein-protein interaction is responsible for (i) catalyzing the formation of the RNAP-promoter closed complex; and (ii) isomerization of the RNAP-promoter complex to the open conformation. CAP's interaction with RNA polymerase causes bending of the DNA near the transcription start site, thus effectively catalyzing the transcription initiation process. CAP's name is derived from its ability to affect transcription of genes involved in many catabolic pathways. For example, when the amount of glucose transported into the cell is low, a cascade of events results in the increase of cytosolic cAMP levels. This increase in cAMP levels is sensed by CAP, which goes on to activate the transcription of many other catabolic genes.
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure.
In molecular genetics, the Krüppel-like family of transcription factors (KLFs) are a set of eukaryotic C2H2 zinc finger DNA-binding proteins that regulate gene expression. This family has been expanded to also include the Sp transcription factor and related proteins, forming the Sp/KLF family.

The trp operon is an operon-a group of genes that is used, or transcribed, together—that codes for the components for production of tryptophan. The trp operon is present in many bacteria, but was first characterized in Escherichia coli. The operon is regulated so that, when tryptophan is present in the environment, the genes for tryptophan synthesis are not expressed. It was an important experimental system for learning about gene regulation, and is commonly used to teach gene regulation.

Directionality, in molecular biology and biochemistry, is the end-to-end chemical orientation of a single strand of nucleic acid. In a single strand of DNA or RNA, the chemical convention of naming carbon atoms in the nucleotide pentose-sugar-ring means that there will be a 5′ end, which frequently contains a phosphate group attached to the 5′ carbon of the ribose ring, and a 3′ end, which typically is unmodified from the ribose -OH substituent. In a DNA double helix, the strands run in opposite directions to permit base pairing between them, which is essential for replication or transcription of the encoded information.
Therapeutic gene modulation refers to the practice of altering the expression of a gene at one of various stages, with a view to alleviate some form of ailment. It differs from gene therapy in that gene modulation seeks to alter the expression of an endogenous gene whereas gene therapy concerns the introduction of a gene whose product aids the recipient directly.

Artificial transcription factors (ATFs) are engineered individual or multi molecule transcription factors that either activate or repress gene transcription (biology).
In a zinc finger protein, certain sequences of amino acid residues are able to recognise and bind to an extended target-site of four or even five nucleotides When this occurs in a ZFP in which the three-nucleotide subsites are contiguous, one zinc finger interferes with the target-site of the zinc finger adjacent to it, a situation known as target-site overlap. For example, a zinc finger containing arginine at position -1 and aspartic acid at position 2 along its alpha-helix will recognise an extended sequence of four nucleotides of the sequence 5'-NNG(G/T)-3'. The hydrogen bond between Asp2 and the N4 of either a cytosine or adenine base paired to the guanine or thymine, respectively defines these two nucleotides at the 3' position, defining a sequence that overlaps into the subsite of any zinc finger that may be attached N-terminally.
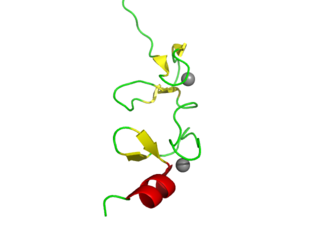
LIM domains are protein structural domains, composed of two contiguous zinc fingers, separated by a two-amino acid residue hydrophobic linker. The domain name is an acronym of the three genes in which it was first identified. LIM is a protein interaction domain that is involved in binding to many structurally and functionally diverse partners. The LIM domain appeared in eukaryotes sometime prior to the most recent common ancestor of plants, fungi, amoeba and animals. In animal cells, LIM domain-containing proteins often shuttle between the cell nucleus where they can regulate gene expression, and the cytoplasm where they are usually associated with actin cytoskeletal structures involved in connecting cells together and to the surrounding matrix, such as stress fibers, focal adhesions and adherens junctions.
Zinc finger protein chimera are chimeric proteins composed of a DNA-binding zinc finger protein domain and another domain through which the protein exerts its effect. The effector domain may be a transcriptional activator (A) or repressor (R), a methylation domain (M) or a nuclease (N).
The RNA-binding Proteins Database (RBPDB) is a biological database of RNA-binding protein specificities that includes experimental observations of RNA-binding sites. The experimental results included are both in vitro and in vivo from primary literature. It includes four metazoan species, which are Homo sapiens, Mus musculus, Drosophila melanogaster, and Caenorhabditis elegans. RNA-binding domains included in this database are RNA recognition motif, K homology, CCCH zinc finger, and more domains. As of 2021, the latest RBPDB release includes 1,171 RNA-binding proteins.

The WRKY domain is found in the WRKY transcription factor family, a class of transcription factors. The WRKY domain is found almost exclusively in plants although WRKY genes appear present in some diplomonads, social amoebae and other amoebozoa, and fungi incertae sedis. They appear absent in other non-plant species. WRKY transcription factors have been a significant area of plant research for the past 20 years. The WRKY DNA-binding domain recognizes the W-box (T)TGAC(C/T) cis-regulatory element.
Transmembrane protein 251, also known as C14orf109 or UPF0694, is a protein that in humans is encoded by the TMEM251 gene. One notable feature of this protein is the presence of proline residues on one of its predicted transmembrane domains., which is a determinant of the intramitochondrial sorting of inner membrane proteins.

Zinc Finger Protein 548 (ZNF548) is a human protein encoded by the ZNF548 gene which is located on chromosome 19. It is found in the nucleus and is hypothesized to play a role in the regulation of transcription by RNA Polymerase II. It belongs to the Krüppel C2H2-type zinc-finger protein family as it contains many zinc-finger repeats.
References
- 1 2 3 Lundin, M.; Nehlin, J. O.; Ronne, H. (1994-03-01). "Importance of a flanking AT-rich region in target site recognition by the GC box-binding zinc finger protein MIG1". Molecular and Cellular Biology. 14 (3): 1979–1985. doi:10.1128/MCB.14.3.1979. ISSN 0270-7306. PMC 358557 . PMID 8114729.
- ↑ Klug WS, Cummings MR, Spencer CA, Palladina MA (2009). Concepts of Genetics (Ninth ed.). San Francisco: Pearson Benjamin Cummings. pp. 463–464. ISBN 978-0-321-54098-0.