
Pseudomonas is a genus of Gram-negative, Gammaproteobacteria, belonging to the family Pseudomonadaceae and containing 191 validly described species. The members of the genus demonstrate a great deal of metabolic diversity and consequently are able to colonize a wide range of niches. Their ease of culture in vitro and availability of an increasing number of Pseudomonas strain genome sequences has made the genus an excellent focus for scientific research; the best studied species include P. aeruginosa in its role as an opportunistic human pathogen, the plant pathogen P. syringae, the soil bacterium P. putida, and the plant growth-promoting P. fluorescens, P. lini, P. migulae, and P. graminis.
Pathogenicity islands (PAIs), as termed in 1990, are a distinct class of genomic islands acquired by microorganisms through horizontal gene transfer. Pathogenicity islands are found in both animal and plant pathogens. Additionally, PAIs are found in both gram-positive and gram-negative bacteria. They are transferred through horizontal gene transfer events such as transfer by a plasmid, phage, or conjugative transposon. Therefore, PAIs contribute to microorganisms' ability to evolve.

Burkholderia is a genus of Proteobacteria whose pathogenic members include the Burkholderia cepacia complex, which attacks humans and Burkholderia mallei, responsible for glanders, a disease that occurs mostly in horses and related animals; Burkholderia pseudomallei, causative agent of melioidosis; and Burkholderia cepacia, an important pathogen of pulmonary infections in people with cystic fibrosis (CF).

Pseudomonas aeruginosa is a common encapsulated, Gram-negative, rod-shaped bacterium that can cause disease in plants and animals, including humans. A species of considerable medical importance, P. aeruginosa is a multidrug resistant pathogen recognized for its ubiquity, its intrinsically advanced antibiotic resistance mechanisms, and its association with serious illnesses – hospital-acquired infections such as ventilator-associated pneumonia and various sepsis syndromes.
The gene-for-gene relationship was discovered by Harold Henry Flor who was working with rust of flax. Flor showed that the inheritance of both resistance in the host and parasite ability to cause disease is controlled by pairs of matching genes. One is a plant gene called the resistance (R) gene. The other is a parasite gene called the avirulence (Avr) gene. Plants producing a specific R gene product are resistant towards a pathogen that produces the corresponding Avr gene product. Gene-for-gene relationships are a widespread and very important aspect of plant disease resistance. Another example can be seen with Lactuca serriola versus Bremia lactucae.

Pseudomonas syringae is a rod-shaped, Gram-negative bacterium with polar flagella. As a plant pathogen, it can infect a wide range of species, and exists as over 50 different pathovars, all of which are available to researchers from international culture collections such as the NCPPB, ICMP, and others.
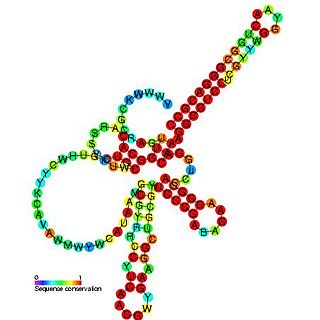
Pseudomonas sRNA P11 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen Pseudomonas aeruginosa and its expression verified by northern blot analysis. P11 is located between a putative threonine protein kinase and putative nitrate reductase and is conserved in several Pseudomonas species. P11 has a predicted Rho independent terminator at the 3′ end but the function of P11 is unknown.
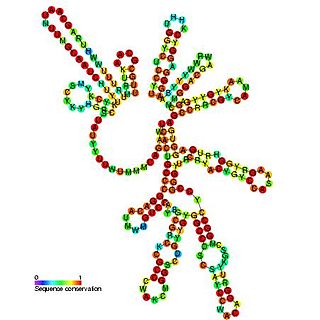
Pseudomonas sRNA P24 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen Pseudomonas aeruginosa and its expression verified by northern blot analysis.
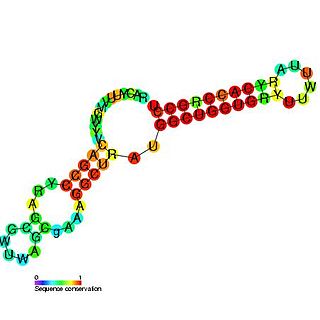
Pseudomonas sRNA P26 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen Pseudomonas aeruginosa and its expression verified by northern blot analysis. P26 is conserved across many Gammaproteobacteria species and appears to be consistently located between the DNA directed RNA polymerase and 50S ribosomal protein L7/L12 genes.
The gamma-150 RNA motif is a conserved RNA structure that is found in bacteria within the order Pseudomonadales. Because gamma-150 RNAs are not consistently in 5' UTRs, the gamma-150 motif is presumed to correspond to a non-coding RNA.

Tabtoxin, also known as wildfire toxin, is a simple monobactam phytotoxin produced by Pseudomonas syringae. It is the precursor to the antibiotic tabtoxinine β-lactam. Tabtoxin is a monocyclic β-lactam produced by P. syringae pv. tabaci, coronafaciens, and garcae. Pseudomonas syringae pv. tabaci, the causal agent of the wildfire of tobacco, produces the phytotoxin tabtoxin. tabtoxin-producing bacterium, P. syringae BR2, causes a disease of bean similar to tobacco wildfire. This organism is closely related to P. syringae pv. tabaci but cannot be classified in the pathovar tabaci because it is not pathogenic on tobacco. Tabtoxin has been shown to be a dipeptide precursor that must undergo hydrolysis by a peptidase to yield the biologically active form, tabtoxinine-p-lactam (TβL). Tabtoxin is required by BR2(R) for both chlorosis and lesion formation on bean. All mutations that affected tabtoxin production, whether spontaneous deletion or transposon induced, also affected lesion formation, and in all cases, restoration of tabtoxin production also restored pathogenic symptoms. Other factors may be required for BR2 to be pathogenic on bean, but apparently these are in addition to tabtoxin production.

The gabT RNA motif is the name of a conserved RNA structure identified by bioinformatics whose function is unknown. The gabT motif has been detected exclusively in bacteria within the genus Pseudomonas, and is found only upstream of gabT genes, and downstream to gabD genes.

The L17 downstream element RNA motif is a conserved RNA structure identified in bacteria by bioinformatics. All known L17 downstream elements were detected immediately downstream of genes encoding the L17 subunit of the ribosome, and therefore might be in the 3' untranslated regions of these genes. The element is found in a variety of lactic acid bacteria and in the genus Listeria.

The livK RNA motif describes a conserved RNA structured that was discovered using bioinformatics. The livK motif is detected only in the species Pseudomonas syringae. It is found in the potential 5' untranslated regions of livK genes and downstream livM and livH genes, as well as the 5' UTRs of amidase genes. The liv genes are predicted to be transporters of branched-chain amino acids, i.e., leucine, isoleucine or valine. The specific reaction catalyzed the amidase genes is not predicted.

The Pseudomon-1 RNA motif is a conserved RNA identified by bioinformatics. It is used by most species whose genomes have been sequenced and that are classified within the genus Pseudomonas, and is also present in Azotobacter vinelandii, a closely related species. It is presumed to function as a non-coding RNA. Pseudomon-1 RNAs consistently have a downstream rho-independent transcription terminator.
The Pseudomon-groES RNA motif is a conserved RNA structure identified in certain bacteria using bioinformatics. It is found in most species within the family Pseudomonadaceae, and is consistently located in the 5' untranslated regions of operons that contain groES genes. RNA transcripts of the groES genes in Pseudomonas aeruginosa where shown experimentally to be initiated at one of two start sites, from promoters called "P1" and "P2". The Pseudomon-groES RNA is in the 5' UTR of transcripts initiated from the P1 site, but is truncated in P2 transcripts. groES genes are involved in the cellular response to heat shock, but it is not thought that the Pseudomonas-groES RNA motif is involved in heat shock regulation. However, it is thought that the motif might regulate groES genes in response to other stimuli.

The Pseudomon-Rho RNA motif refers to a conserved RNA structure that was discovered using bioinformatics. The RNAs that conform to this motif are found in species within the genus Pseudomonas, as well as the related Azotobacter vinelandii. They are consistently located in what could be the 5' untranslated regions of genes that encode the Rho factor protein, and this arrangement in bacteria suggested that Pseudomon-Rho RNAs might be cis-regulatory elements that regulate concentrations of the Rho protein.

The rmf RNA motif is a conserved RNA structure that was originally detected using bioinformatics. rmf RNAs are consistently foundwithin species classified into the genus Pseudomonas, and is located potentially in the 5′ untranslated regions of rmf genes. These genes encodes the ribosome modulation factor protein, which affects the translation of genes by modifying ribosome structure in response to stress such as starvation. This ribosome modulation is a part of the stringent response in bacteria. The likely biological role of rmf RNAs is ambiguous. Since the RNA could be in the 5′ UTRs of protein-coding genes, it was hypothesized that it functions as a cis-regulatory element. This hypothesis is bolstered by the observation that ribosome modulation factor binds ribosomal RNA, and many cis-regulatory RNAs called ribosomal protein leaders participate in a feedback regulation mechanism by binding to proteins that normally bind to ribosomal RNA. However, since rmf RNAs are not very close to the rmf genes, they might function as non-coding RNAs.
Bacterial small RNAs (sRNA) are small RNAs produced by bacteria; they are 50- to 500-nucleotide non-coding RNA molecules, highly structured and containing several stem-loops. Numerous sRNAs have been identified using both computational analysis and laboratory-based techniques such as Northern blotting, microarrays and RNA-Seq in a number of bacterial species including Escherichia coli, the model pathogen Salmonella, the nitrogen-fixing alphaproteobacterium Sinorhizobium meliloti, marine cyanobacteria, Francisella tularensis, Streptococcus pyogenes, the pathogen Staphylococcus aureus, and the plant pathogen Xanthomonas oryzae pathovar oryzae. Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNA or protein, and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress response, and structure.

The ivy-DE RNA motif is a conserved RNA structure that was discovered by bioinformatics. ivy-DE motifs are found in the genus Pseudomonas.















