Hepatitis D is a type of viral hepatitis caused by the hepatitis delta virus (HDV). HDV is one of five known hepatitis viruses: A, B, C, D, and E. HDV is considered to be a satellite because it can propagate only in the presence of the hepatitis B virus (HBV). Transmission of HDV can occur either via simultaneous infection with HBV (coinfection) or superimposed on chronic hepatitis B or hepatitis B carrier state (superinfection).

Hepadnaviridae is a family of viruses. Humans, apes, and birds serve as natural hosts. There are currently 18 species in this family, divided among 5 genera. Its best-known member is hepatitis B virus. Diseases associated with this family include: liver infections, such as hepatitis, hepatocellular carcinomas, and cirrhosis. It is the sole accepted family in the order Blubervirales.
Duck hepatitis B virus, abbreviated DHBV, is part of the genus Avihepadnavirus of the Hepadnaviridae, and is the causal agent of duck hepatitis B.
The murine leukemia viruses are retroviruses named for their ability to cause cancer in murine (mouse) hosts. Some MLVs may infect other vertebrates. MLVs include both exogenous and endogenous viruses. Replicating MLVs have a positive sense, single-stranded RNA (ssRNA) genome that replicates through a DNA intermediate via the process of reverse transcription.

This family represents the bovine leukaemia virus RNA encapsidation (packaging) signal, which is essential for efficient viral replication.

The Citrus tristeza virus replication signal is a regulatory element involved in a viral replication signal which is highly conserved in citrus tristeza viruses. Replication signals are required for viral replication and are usually found near the 5' and 3' termini of protein coding genes. This element is predicted to form ten stem loop structures some of which are essential for functions that provide for efficient viral replication.

The Coronavirus packaging signal is a conserved cis-regulatory element found in Betacoronavirus. It has an important role in regulating the packaging of the viral genome into the capsid. As part of the viral life cycle, within the infected cell, the viral genome becomes associated with viral proteins and assembles into new infective progeny viruses. This process is called packaging and is vital for viral replication.

The Gammaretrovirus core encapsidation signal is an RNA element known to be essential for stable dimerisation and efficient genome packaging during virus assembly. Dimerisation of the viral RNA genomes is proposed to act as an RNA conformational switch which exposes conserved UCUG elements and enables efficient genome encapsidation. The structure of this element is composed of three stem-loops. Two of the stem-loops called SL-C and SL-D form a single co-axial extend helix.

The hepatitis C virus 3′X element is an RNA element which contains three stem-loop structures that are essential for replication.

The retroviral psi packaging element, also known as the Ψ RNA packaging signal, is a cis-acting RNA element identified in the genomes of the retroviruses Human immunodeficiency virus (HIV) and Simian immunodeficiency virus (SIV). It is involved in regulating the essential process of packaging the retroviral RNA genome into the viral capsid during replication. The final virion contains a dimer of two identical unspliced copies of the viral genome.
The Hepatitis B virus PRE stem-loop alpha is an RNA structure that is shown to play a role in nuclear export of HBV mRNAs.
The Hepatitis B virus PRE stem-loop beta is an RNA structure that is shown to play a role in nuclear export of HBV mRNAs.
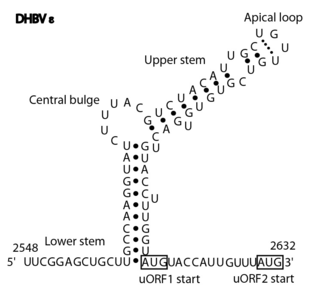
The Avian HBV RNA encapsidation signal epsilon is an RNA structure that is shown to facilitate encapsidation of the pregenomic RNA required for viral replication. There are two main classes of encapsidation signals in avian hepatitis B viruses - Duck hepatitis B virus (DHBV) and Heron hepatitis B virus (HHBV) like. DHBV is used as a model to understand human Hepatitis B virus. Although studies have shown that the HHBV epsilon has less pairing in the upper stem than DHBV, this pairing is not absolutely required for DHBV infection in ducks.

HBx is a hepatitis B viral protein. It is 154 amino acids long and interferes with transcription, signal transduction, cell cycle progress, protein degradation, apoptosis and chromosomal stability in the host. It forms a heterodimeric complex with its cellular target protein, and this interaction dysregulates centrosome dynamics and mitotic spindle formation. It interacts with DDB1 redirecting the ubiquitin ligase activity of the CUL4-DDB1 E3 complexes, which are intimately involved in the intracellular regulation of DNA replication and repair, transcription and signal transduction.

Hepatitis B virus DNA polymerase is a hepatitis B viral protein. It is a DNA polymerase that can use either DNA or RNA templates and a ribonuclease H that cuts RNA in the duplex. Both functions are supplied by the reverse transcriptase (RT) domain.

Hepatitis B virus (HBV) is a partially double-stranded DNA virus, a species of the genus Orthohepadnavirus and a member of the Hepadnaviridae family of viruses. This virus causes the disease hepatitis B.
The transmission of hepadnaviruses between their natural hosts, humans, non-human primates, and birds, including intra-species host transmission and cross-species transmission, is a topic of study in virology.
Hepatitis B virus PRE 1151–1410 is a part of 500 base pair long HBV PRE, that has been proposed to be the hepatitis B virus (HBV) RNA export element. However, the function is controversial and new regulatory elements have been predicted within PRE. PRE 1151–1410 enhances nuclear export of intronless transcripts and represses the splicing mechanism to a comparable degree to that of the full-length PRE. Hence it was proposed to be the core HBV PRE element. PRE1151–1410 contains 3 known regulatory elements: PRE SL-alpha, human La protein binding site, SRE-1.

Ground squirrel hepatitis virus, abbreviated GSHV, is a partially double-stranded DNA virus that is closely related to human Hepatitis B virus (HBV) and Woodchuck hepatitis virus (WHV). It is a member of the family of viruses Hepadnaviridae and the genus Orthohepadnavirus. Like the other members of its family, GSHV has high degree of species and tissue specificity. It was discovered in Beechey ground squirrels, Spermophilus beecheyi, but also infects Arctic ground squirrels, Spermophilus parryi. Commonalities between GSHV and HBV include morphology, DNA polymerase activity in genome repair, cross-reacting viral antigens, and the resulting persistent infection with viral antigen in the blood (antigenemia). As a result, GSHV is used as an experimental model for HBV.

The woolly monkey hepatitis B virus (WMHBV) is a viral species of the Orthohepadnavirus genus of the Hepadnaviridae family. Its natural host is the woolly monkey (Lagothrix), an inhabitant of South America categorized as a New World primate. WMHBV, like other hepatitis viruses, infects the hepatocytes, or liver cells, of its host organism. It can cause hepatitis, liver necrosis, cirrhosis, and hepatocellular carcinoma. Because nearly all species of Lagothrix are threatened or endangered, researching and developing a vaccine and/or treatment for WMHBV is important for the protection of the whole woolly monkey genus.












