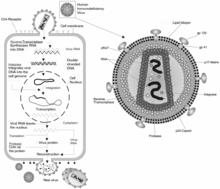
A retrovirus is a type of virus that inserts a DNA copy of its RNA genome into the DNA of a host cell that it invades, thus changing the genome of that cell. Once inside the host cell's cytoplasm, the virus uses its own reverse transcriptase enzyme to produce DNA from its RNA genome, the reverse of the usual pattern, thus retro (backwards). The new DNA is then incorporated into the host cell genome by an integrase enzyme, at which point the retroviral DNA is referred to as a provirus. The host cell then treats the viral DNA as part of its own genome, transcribing and translating the viral genes along with the cell's own genes, producing the proteins required to assemble new copies of the virus. Many retroviruses cause serious diseases in humans, other mammals, and birds.

A reverse transcriptase (RT) is an enzyme used to generate complementary DNA (cDNA) from an RNA template, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobile genetic elements to proliferate within the host genome, and by eukaryotic cells to extend the telomeres at the ends of their linear chromosomes. Contrary to a widely held belief, the process does not violate the flows of genetic information as described by the classical central dogma, as transfers of information from RNA to DNA are explicitly held possible.

Retroviral integrase (IN) is an enzyme produced by a retrovirus that integrates—forms covalent links between—its genetic information into that of the host cell it infects. Retroviral INs are not to be confused with phage integrases (recombinases) used in biotechnology, such as λ phage integrase, as discussed in site-specific recombination.

Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules called non-coding RNAs (ncRNAs). mRNA comprises only 1–3% of total RNA samples. Less than 2% of the human genome can be transcribed into mRNA, while at least 80% of mammalian genomic DNA can be actively transcribed, with the majority of this 80% considered to be ncRNA.

Ribonuclease H is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes.
A transposase is any of a class of enzymes capable of binding to the end of a transposon and catalysing its movement to another part of a genome, typically by a cut-and-paste mechanism or a replicative mechanism, in a process known as transposition. The word "transposase" was first coined by the individuals who cloned the enzyme required for transposition of the Tn3 transposon. The existence of transposons was postulated in the late 1940s by Barbara McClintock, who was studying the inheritance of maize, but the actual molecular basis for transposition was described by later groups. McClintock discovered that some segments of chromosomes changed their position, jumping between different loci or from one chromosome to another. The repositioning of these transposons allowed other genes for pigment to be expressed. Transposition in maize causes changes in color; however, in other organisms, such as bacteria, it can cause antibiotic resistance. Transposition is also important in creating genetic diversity within species and generating adaptability to changing living conditions.
Deoxyribozymes, also called DNA enzymes, DNAzymes, or catalytic DNA, are DNA oligonucleotides that are capable of performing a specific chemical reaction, often but not always catalytic. This is similar to the action of other biological enzymes, such as proteins or ribozymes . However, in contrast to the abundance of protein enzymes in biological systems and the discovery of biological ribozymes in the 1980s, there is only little evidence for naturally occurring deoxyribozymes. Deoxyribozymes should not be confused with DNA aptamers which are oligonucleotides that selectively bind a target ligand, but do not catalyze a subsequent chemical reaction.
Lentivirus is a genus of retroviruses that cause chronic and deadly diseases characterized by long incubation periods, in humans and other mammalian species. The genus includes the human immunodeficiency virus (HIV), which causes AIDS. Lentiviruses are distributed worldwide, and are known to be hosted in apes, cows, goats, horses, cats, and sheep as well as several other mammals.
The genome and proteins of HIV (human immunodeficiency virus) have been the subject of extensive research since the discovery of the virus in 1983. "In the search for the causative agent, it was initially believed that the virus was a form of the Human T-cell leukemia virus (HTLV), which was known at the time to affect the human immune system and cause certain leukemias. However, researchers at the Pasteur Institute in Paris isolated a previously unknown and genetically distinct retrovirus in patients with AIDS which was later named HIV." Each virion comprises a viral envelope and associated matrix enclosing a capsid, which itself encloses two copies of the single-stranded RNA genome and several enzymes. The discovery of the virus itself occurred two years following the report of the first major cases of AIDS-associated illnesses.
Cre-Lox recombination is a site-specific recombinase technology, used to carry out deletions, insertions, translocations and inversions at specific sites in the DNA of cells. It allows the DNA modification to be targeted to a specific cell type or be triggered by a specific external stimulus. It is implemented both in eukaryotic and prokaryotic systems. The Cre-lox recombination system has been particularly useful to help neuroscientists to study the brain in which complex cell types and neural circuits come together to generate cognition and behaviors. NIH Blueprint for Neuroscience Research has created several hundreds of Cre driver mouse lines which are currently used by the worldwide neuroscience community.
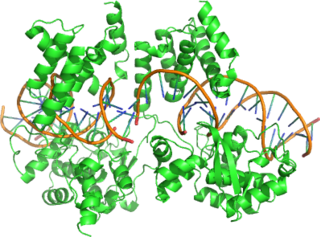
Cre recombinase is a tyrosine recombinase enzyme derived from the P1 bacteriophage. The enzyme uses a topoisomerase I-like mechanism to carry out site specific recombination events. The enzyme (38kDa) is a member of the integrase family of site specific recombinase and it is known to catalyse the site specific recombination event between two DNA recognition sites. This 34 base pair (bp) loxP recognition site consists of two 13 bp palindromic sequences which flank an 8bp spacer region. The products of Cre-mediated recombination at loxP sites are dependent upon the location and relative orientation of the loxP sites. Two separate DNA species both containing loxP sites can undergo fusion as the result of Cre mediated recombination. DNA sequences found between two loxP sites are said to be "floxed". In this case the products of Cre mediated recombination depends upon the orientation of the loxP sites. DNA found between two loxP sites oriented in the same direction will be excised as a circular loop of DNA whilst intervening DNA between two loxP sites that are opposingly orientated will be inverted. The enzyme requires no additional cofactors or accessory proteins for its function.

APOBEC3G is a human enzyme encoded by the APOBEC3G gene that belongs to the APOBEC superfamily of proteins. This family of proteins has been suggested to play an important role in innate anti-viral immunity. APOBEC3G belongs to the family of cytidine deaminases that catalyze the deamination of cytidine to uridine in the single stranded DNA substrate. The C-terminal domain of A3G renders catalytic activity, several NMR and crystal structures explain the substrate specificity and catalytic activity.
Site-specific recombination, also known as conservative site-specific recombination, is a type of genetic recombination in which DNA strand exchange takes place between segments possessing at least a certain degree of sequence homology. Enzymes known as site-specific recombinases (SSRs) perform rearrangements of DNA segments by recognizing and binding to short, specific DNA sequences (sites), at which they cleave the DNA backbone, exchange the two DNA helices involved, and rejoin the DNA strands. In some cases the presence of a recombinase enzyme and the recombination sites is sufficient for the reaction to proceed; in other systems a number of accessory proteins and/or accessory sites are required. Many different genome modification strategies, among these recombinase-mediated cassette exchange (RMCE), an advanced approach for the targeted introduction of transcription units into predetermined genomic loci, rely on SSRs.

A resistance mutation is a mutation in a virus gene that allows the virus to become resistant to treatment with a particular antiviral drug. The term was first used in the management of HIV, the first virus in which genome sequencing was routinely used to look for drug resistance. At the time of infection, a virus will infect and begin to replicate within a preliminary cell. As subsequent cells are infected, random mutations will occur in the viral genome. When these mutations begin to accumulate, antiviral methods will kill the wild type strain, but will not be able to kill one or many mutated forms of the original virus. At this point a resistance mutation has occurred because the new strain of virus is now resistant to the antiviral treatment that would have killed the original virus. Resistance mutations are evident and widely studied in HIV due to its high rate of mutation and prevalence in the general population. Resistance mutation is now studied in bacteriology and parasitology.

HIV-1 protease (PR) is a retroviral aspartyl protease (retropepsin), an enzyme involved with peptide bond hydrolysis in retroviruses, that is essential for the life-cycle of HIV, the retrovirus that causes AIDS. HIV protease cleaves newly synthesized polyproteins at nine cleavage sites to create the mature protein components of an HIV virion, the infectious form of a virus outside of the host cell. Without effective HIV protease, HIV virions remain uninfectious.

PC4 and SFRS1 interacting protein 1, also known as lens epithelium-derived growth factor (LEDGF/p75), dense fine speckles 70kD protein or transcriptional coactivator p75/p52, is a protein that in humans is encoded by the PSIP1 gene.
The pre-integration complex (PIC) is a nucleoprotein complex of viral genetic material and associated viral and host proteins which is capable of inserting a viral genome into a host genome. The PIC forms after uncoating of a viral particle after entry into the host cell. In the case of the human immunodeficiency virus (HIV), the PIC forms after the Reverse Transcription Complex (RTC) has reverse transcribed the viral RNA into DNA. The PIC consists of viral proteins, host proteins and the viral DNA. The PIC enters the cellular nucleus through the nuclear pore complex without disrupting the nuclear envelope, thus allowing HIV and related retroviruses to replicate in non-dividing cells. Following nuclear entry, the PIC's DNA payload may be integrated into the host DNA as a "provirus".

Integrasone is a polyketide natural product, isolated from an unknown fungus, that has been shown to inhibit the HIV-1 integrase enzyme.
The first human immunodeficiency virus (HIV) case was reported in the United States in the early 1980s. Many drugs have been discovered to treat the disease but mutations in the virus and resistance to the drugs make development difficult. Integrase is a viral enzyme that integrates retroviral DNA into the host cell genome. Integrase inhibitors are a new class of drugs used in the treatment of HIV. The first integrase inhibitor, raltegravir, was approved in 2007 and other drugs were in clinical trials in 2011.
Daria J Hazuda is a biochemist known for discovering the first HIV Integrase Strand Transfer Inhibitors (InSTIs) and leading the development of the first HIV integrase inhibitor to gain FDA approval, Isentress (raltegravir). Her lab also determined these inhibitors' mechanism of action and ways the virus could develop resistance to them. Hazuda has also performed extensive research and led the development of antivirals for Hepatitis C Virus (HCV) including Elbasvir and Grazoprevir. She currently serves as Vice President for Infectious Diseases Discovery for Merck and Chief Scientific Officer of their MRL Cambridge Exploratory Science Center.