Proteins are large biomolecules, or macromolecules, consisting of one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells, and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity.

A sarcomere is the complicated unit of striated muscle tissue. It is the repeating unit between two Z lines. Skeletal muscles are composed of tubular muscle cells which are formed in a process known as myogenesis. Muscle fibers contain numerous tubular myofibrils. Myofibrils are composed of repeating sections of sarcomeres, which appear under the microscope as alternating dark and light bands. Sarcomeres are composed of long, fibrous proteins as filaments that slide past each other when a muscle contracts or relaxes. The costamere is a different component that connects the sarcomere to the sarcolemma.

Har Gobind Khorana was an Indian American biochemist. While on the faculty of the University of Wisconsin–Madison, he shared the 1968 Nobel Prize for Physiology or Medicine with Marshall W. Nirenberg and Robert W. Holley for research that showed the order of nucleotides in nucleic acids, which carry the genetic code of the cell and control the cell's synthesis of proteins. Khorana and Nirenberg were also awarded the Louisa Gross Horwitz Prize from Columbia University in the same year.
Vamp or vamps may refer to:
An oligomer is a molecular complex of chemicals that consists of a few repeating units, in contrast to a polymer, where the number of monomers is, in principle, infinite. Dimers, trimers, and tetramers are, for instance, oligomers composed of two, three, and four monomers, respectively.

Glycosaminoglycans (GAGs) or mucopolysaccharides are long linear polysaccharides consisting of repeating disaccharide units. Except for keratan, the repeating unit consists of an amino sugar, along with a uronic sugar or galactose. Because GAGs are highly polar and attract water, they are used in the body as a lubricant or shock absorber.
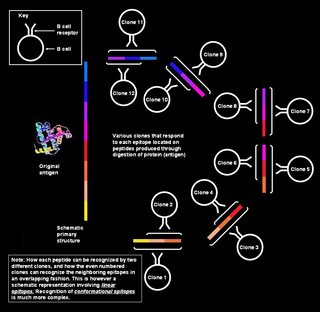
A linear or a sequential epitope is an epitope that is recognized by antibodies by its linear sequence of amino acids, or primary structure. In contrast, most antibodies recognize a conformational epitope that has a specific three-dimensional shape and its protein structure.
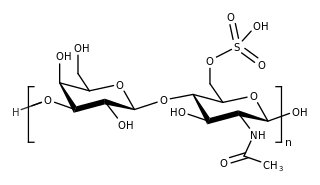
Keratan sulfate (KS), also called keratosulfate, is any of several sulfated glycosaminoglycans that have been found especially in the cornea, cartilage, and bone. It is also synthesized in the central nervous system where it participates both in development and in the glial scar formation following an injury. Keratan sulfates are large, highly hydrated molecules which in joints can act as a cushion to absorb mechanical shock.
The Protein Data Bank (pdb) file format is a textual file format describing the three-dimensional structures of molecules held in the Protein Data Bank. The pdb format accordingly provides for description and annotation of protein and nucleic acid structures including atomic coordinates, secondary structure assignments, as well as atomic connectivity. In addition experimental metadata are stored. PDB format is the legacy file format for the Protein Data Bank which now keeps data on biological macromolecules in the newer mmCIF file format.
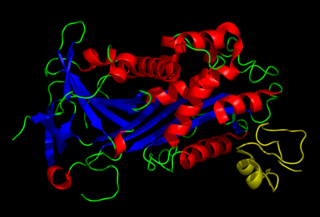
Vitronectin is a glycoprotein of the hemopexin family which is abundantly found in serum, the extracellular matrix and bone. In humans it is encoded by the VTN gene.

A pi helix is a type of secondary structure found in proteins. Discovered by crystallographer Barbara Low in 1952 and once thought to be rare, short π-helices are found in 15% of known protein structures and are believed to be an evolutionary adaptation derived by the insertion of a single amino acid into an α-helix. Because such insertions are highly destabilizing, the formation of π-helices would tend to be selected against unless it provided some functional advantage to the protein. π-helices therefore are typically found near functional sites of proteins.

An alpha solenoid is a protein fold composed of repeating alpha helix subunits, commonly helix-turn-helix motifs, arranged in antiparallel fashion to form a superhelix. Alpha solenoids are known for their flexibility and plasticity. Like beta propellers, alpha solenoids are a form of solenoid protein domain commonly found in the proteins comprising the nuclear pore complex. They are also common in membrane coat proteins known as coatomers, such as clathrin, and in regulatory proteins that form extensive protein-protein interactions with their binding partners. Examples of alpha solenoid structures binding RNA and lipids have also been described.
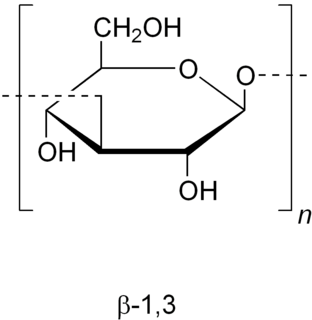
Zymosan is a glucan with repeating glucose units connected by β-1,3-glycosidic linkages. It binds to TLR 2 and Dectin-1 (CLEC7A). Zymosan is a ligand found on the surface of fungi, like yeast.
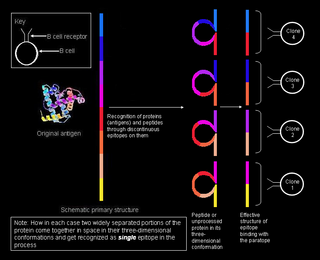
A conformational epitope is a sequence of sub-units composing an antigen that come in direct contact with a receptor of the immune system.

The WD40 repeat is a short structural motif of approximately 40 amino acids, often terminating in a tryptophan-aspartic acid (W-D) dipeptide. Tandem copies of these repeats typically fold together to form a type of circular solenoid protein domain called the WD40 domain.
The Unique Ingredient Identifier (UNII) is a non-proprietary, free, unique, unambiguous, non-semantic, alphanumeric identifier linked to a substance's molecular structure or descriptive information and generated by the Global Substance Registration System (GSRS) of the Food and Drug Administration (FDA).
O-linked glycosylation is the attachment of a sugar molecule to the oxygen atom of serine (Ser) or threonine (Thr) residues in a protein. O-glycosylation is a post-translational modification that occurs after the protein has been synthesised. In eukaryotes, it occurs in the endoplasmic reticulum, Golgi apparatus and occasionally in the cytoplasm; in prokaryotes, it occurs in the cytoplasm. Several different sugars can be added to the serine or threonine, and they affect the protein in different ways by changing protein stability and regulating protein activity. O-glycans, which are the sugars added to the serine or threonine, have numerous functions throughout the body, including trafficking of cells in the immune system, allowing recognition of foreign material, controlling cell metabolism and providing cartilage and tendon flexibility. Because of the many functions they have, changes in O-glycosylation are important in many diseases including cancer, diabetes and Alzheimer's. O-glycosylation occurs in all domains of life, including eukaryotes, archaea and a number of pathogenic bacteria including Burkholderia cenocepacia, Neisseria gonorrhoeae and Acinetobacter baumannii.

The term macromolecular assembly (MA) refers to massive chemical structures such as viruses and non-biologic nanoparticles, cellular organelles and membranes and ribosomes, etc. that are complex mixtures of polypeptide, polynucleotide, polysaccharide or other polymeric macromolecules. They are generally of more than one of these types, and the mixtures are defined spatially, and with regard to their underlying chemical composition and structure. Macromolecules are found in living and nonliving things, and are composed of many hundreds or thousands of atoms held together by covalent bonds; they are often characterized by repeating units. Assemblies of these can likewise be biologic or non-biologic, though the MA term is more commonly applied in biology, and the term supramolecular assembly is more often applied in non-biologic contexts. MAs of macromolecules are held in their defined forms by non-covalent intermolecular interactions, and can be in either non-repeating structures, or in repeating linear, circular, spiral, or other patterns. The process by which MAs are formed has been termed molecular self-assembly, a term especially applied in non-biologic contexts. A wide variety of physical/biophysical, chemical/biochemical, and computational methods exist for the study of MA; given the scale of MAs, efforts to elaborate their composition and structure and discern mechanisms underlying their functions are at the forefront of modern structure science.

Abductin is a natural elastic protein that is found in the hinge ligament of bivalve mollusks. It is unique as it is the only protein in nature with compressible elasticity. It is similar to elastin and resilin, but amino acid analysis reveals that it has high concentrations of glycine and methionine.

The beta bend ribbon, or beta-bend ribbon, is a structural feature in polypeptides and proteins. The shortest possible has six amino acid residues arranged as two overlapping hydrogen bonded beta turns in which the carbonyl group of residue i is hydrogen-bonded to the NH of residue i+3 while the carbonyl group of residue i+2 is hydrogen-bonded to the NH of residue i+5. In longer ribbons, this bonding is continued in peptides of 8, 10, etc., amino acid residues. A beta bend ribbon can be regarded as an aberrant 310 helix (3/10-helix) that has lost some of its hydrogen bonds. Two websites are available to facilitate finding and examining these features in proteins: Motivated Proteins; and PDBeMotif.