Related Research Articles

Microevolution is the change in allele frequencies that occurs over time within a population. This change is due to four different processes: mutation, selection, gene flow and genetic drift. This change happens over a relatively short amount of time compared to the changes termed macroevolution.

A transposable element is a DNA sequence that can change its position within a genome, sometimes creating or reversing mutations and altering the cell's genetic identity and genome size. Transposition often results in duplication of the same genetic material. Barbara McClintock's discovery of them earned her a Nobel Prize in 1983.
Non-coding DNA sequences are components of an organism's DNA that do not encode protein sequences. Some non-coding DNA is transcribed into functional non-coding RNA molecules. Other functions of non-coding DNA include the transcriptional and translational regulation of protein-coding sequences, scaffold attachment regions, origins of DNA replication, centromeres and telomeres. Its RNA counterpart is non-coding RNA.

Horizontal gene transfer (HGT) or lateral gene transfer (LGT) is the movement of genetic material between unicellular and/or multicellular organisms other than by the ("vertical") transmission of DNA from parent to offspring (reproduction). HGT is an important factor in the evolution of many organisms.
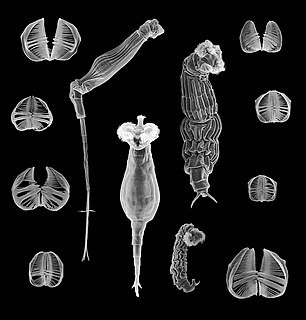
Bdelloidea is a class of rotifers found in freshwater habitats all over the world. There are over 450 described species of bdelloid rotifers, distinguished from each other mainly on the basis of morphology. The main characteristics that distinguish bdelloids from related groups of rotifers are exclusively parthenogenetic reproduction and the ability to survive in dry, harsh environments by entering a state of desiccation-induced dormancy (anhydrobiosis) at any life stage. They are often referred to as "ancient asexuals" due to their unique asexual history that spans back to over 25 million years ago through fossil evidence. Bdelloid rotifers are microscopic organisms, typically between 150 and 700 µm in length. Most are slightly too small to be seen with the naked eye, but appear as tiny white dots through even a weak hand lens, especially in bright light. In June 2021, biologists reported the restoration of bdelloid rotifers after being frozen for 24,000 years in the Siberian permafrost.

A gene family is a set of several similar genes, formed by duplication of a single original gene, and generally with similar biochemical functions. One such family are the genes for human hemoglobin subunits; the ten genes are in two clusters on different chromosomes, called the α-globin and β-globin loci. These two gene clusters are thought to have arisen as a result of a precursor gene being duplicated approximately 500 million years ago.

CRISPR is a family of DNA sequences found in the genomes of prokaryotic organisms such as bacteria and archaea. These sequences are derived from DNA fragments of bacteriophages that had previously infected the prokaryote. They are used to detect and destroy DNA from similar bacteriophages during subsequent infections. Hence these sequences play a key role in the antiviral defense system of prokaryotes and provide a form of acquired immunity. CRISPR are found in approximately 50% of sequenced bacterial genomes and nearly 90% of sequenced archaea.

Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids. It is widely used by cells to accurately repair harmful breaks that occur on both strands of DNA, known as double-strand breaks (DSB), in a process called homologous recombinational repair (HRR). Homologous recombination also produces new combinations of DNA sequences during meiosis, the process by which eukaryotes make gamete cells, like sperm and egg cells in animals. These new combinations of DNA represent genetic variation in offspring, which in turn enables populations to adapt during the course of evolution. Homologous recombination is also used in horizontal gene transfer to exchange genetic material between different strains and species of bacteria and viruses.

In biology, a gene is a basic unit of heredity and a sequence of nucleotides in DNA that encodes the synthesis of a gene product, either RNA or protein.

The Rafflesiaceae are a family of rare parasitic plants comprising 36 species in 3 genera found in the tropical forests of east and southeast Asia, including Rafflesia arnoldii, which has the largest flowers of all plants. The plants are endoparasites of vines in the genus Tetrastigma (Vitaceae) and lack stems, leaves, roots, and any photosynthetic tissue. They rely entirely on their host plants for both water and nutrients, and only then emerge as flowers from the roots or lower stems of the host plants.

Complement receptor type 2 (CR2), also known as complement C3d receptor, Epstein-Barr virus receptor, and CD21, is a protein that in humans is encoded by the CR2 gene.

A prokaryote is a unicellular organism that lacks a nuclear membrane-enclosed nucleus. The word prokaryote comes from the Greek πρό and κάρυον. In the two-empire system arising from the work of Édouard Chatton, prokaryotes were classified within the empire Prokaryota. But in the three-domain system, based upon molecular analysis, prokaryotes are divided into two domains: Bacteria and Archaea. Organisms with nuclei are placed in a third domain, Eukaryota. In the study of the origins of life, prokaryotes are thought to have arisen before eukaryotes.
Concerted evolution is the phenomenon where paralogous genes within one species are more closely related to one another than to members of the same gene family in closely related species. It is possible that this might occur even if the gene duplication event preceded the speciation event. High sequence similarity between paralogs may be maintained by homologous recombination events that lead to gene conversion, effectively copying some sequence from one and overwriting the homologous region in the other. Another possible hypothesis that has yet to be disproved is that rapid waves of gene duplication are responsible for the apparently "concerted" homogeneity of tandem and unlinked repeats seen in concerted evolution.
Helitrons are one of the three groups of eukaryotic class 2 transposable elements (TEs) so far described. They are the eukaryotic rolling-circle transposable elements which are hypothesized to transpose by a rolling circle replication mechanism via a single-stranded DNA intermediate. They were first discovered in plants and in the nematode Caenorhabditis elegans, and now they have been identified in a diverse range of species, from protists to mammals. Helitrons make up a substantial fraction of many genomes where non-autonomous elements frequently outnumber the putative autonomous partner. Helitrons seem to have a major role in the evolution of host genomes. They frequently capture diverse host genes, some of which can evolve into novel host genes or become essential for Helitron transposition.

Gene transfer agents (GTAs) are DNA-containing virus-like particles that are produced by some bacteria and archaea and mediate horizontal gene transfer. Different GTA types have originated independently from viruses in several bacterial and archaeal lineages. These cells produce GTA particles containing short segments of the DNA present in the cell. After the particles are released from the producer cell, they can attach to related cells and inject their DNA into the cytoplasm. The DNA can then become part of the recipient cells' genome.
Incomplete lineage sorting, also termed deep coalescence, retention of ancestral polymorphism, or trans-species polymorphism, describes a phenomenon in population genetics when ancestral gene copies fail to coalesce into a common ancestral copy until deeper than previous speciation events. In other words, the tree produced by a single gene differs from the population or species level tree, producing a discordant tree. Effects caused by lineage sorting of genetic polymorphisms that were retained across successive nodes in the species tree have been called hemiplasy. Whatever the mechanism, the result is that a generated species level tree may differ depending on the selected genes used for assessment. This is in contrast to complete lineage sorting, where the tree produced by the gene is the same as the population or species level tree. Both are common results in phylogenetic analysis, although it depends on the gene, organism, and sampling technique.
Horizontal or lateral gene transfer is the transmission of portions of genomic DNA between organisms through a process decoupled from vertical inheritance. In the presence of HGT events, different fragments of the genome are the result of different evolutionary histories. This can therefore complicate the investigations of evolutionary relatedness of lineages and species. Also, as HGT can bring into genomes radically different genotypes from distant lineages, or even new genes bearing new functions, it is a major source of phenotypic innovation and a mechanism of niche adaptation. For example, of particular relevance to human health is the lateral transfer of antibiotic resistance and pathogenicity determinants, leading to the emergence of pathogenic lineages.
DNA transposons are DNA sequences, sometimes referred to "jumping genes", that can move and integrate to different locations within the genome. They are class II transposable elements (TEs) that move through a DNA intermediate, as opposed to class I TEs, retrotransposons, that move through an RNA intermediate. DNA transposons can move in the DNA of an organism via a single-or double-stranded DNA intermediate. DNA transposons have been found in both prokaryotic and eukaryotic organisms. They can make up a significant portion of an organism's genome, particularly in eukaryotes. In prokaryotes, TE's can facilitate the horizontal transfer of antibiotic resistance or other genes associated with virulence. After replicating and propagating in a host, all transposon copies become inactivated and are lost unless the transposon passes to a genome by starting a new life cycle with horizontal transfer. It is important to note that DNA transposons do not randomly insert themselves into the genome, but rather show preference for specific sites.
Tc1/mariner is a class and superfamily of interspersed repeats DNA transposons. The elements of this class are found in all animals, including humans. They can also be found in protists and bacteria.
hAT transposons are a superfamily of DNA transposons, or Class II transposable elements, that are common in the genomes of plants, animals, and fungi.
References
- ↑ Clemenza, Liliana; Subramanian, Bala; Hourcade, Dennis; Nickells, Michael; Atkinson, John (1997). "Primary sequence of baboon CR1 demonstrates concerted evolution within the CR1 gene". Molecular Immunology. Elsevier BV. 34 (4): 297–304. doi:10.1016/s0161-5890(97)00040-0. ISSN 0161-5890. PMID 9244342.
- ↑ Wolf, Yuri I.; Aravind, L.; Grishin, Nick V.; Koonin, Eugene V. (1999-09-09). "Evolution of Aminoacyl-tRNA Synthetases—Analysis of Unique Domain Architectures and Phylogenetic Trees Reveals a Complex History of Horizontal Gene Transfer Events". Genome Research. 9 (8): 689–710. doi:10.1101/gr.9.8.689 (inactive 31 October 2021). PMID 10447505.CS1 maint: DOI inactive as of October 2021 (link)