
In the fields of molecular biology and genetics, a genome is all genetic information of an organism. It consists of nucleotide sequences of DNA. The nuclear genome includes protein-coding genes and non-coding genes, the other functional regions of the genome, and any junk DNA if it is present. Algae and plants contain chloroplasts with a chloroplast genome and almost all eukaryotes have mitochondria and a mitochondrial genome.

The human genome is a complete set of nucleic acid sequences for humans, encoded as DNA within the 23 chromosome pairs in cell nuclei and in a small DNA molecule found within individual mitochondria. These are usually treated separately as the nuclear genome and the mitochondrial genome. Human genomes include both protein-coding DNA sequences and various types of DNA that does not encode proteins. The latter is a diverse category that includes DNA coding for non-translated RNA, such as that for ribosomal RNA, transfer RNA, ribozymes, small nuclear RNAs, and several types of regulatory RNAs. It also includes promoters and their associated gene-regulatory elements, DNA playing structural and replicatory roles, such as scaffolding regions, telomeres, centromeres, and origins of replication, plus large numbers of transposable elements, inserted viral DNA, non-functional pseudogenes and simple, highly-repetitive sequences. Introns make up a large percentage of non-coding DNA. Some of this non-coding DNA is non-functional junk DNA, such as pseudogenes, but there is no firm consensus on the total mount of junk DNA.
Non-coding DNA (ncDNA) sequences are components of an organism's DNA that do not encode protein sequences. Some non-coding DNA is transcribed into functional non-coding RNA molecules. Other functional regions of the non-coding DNA fraction include regulatory sequences that control gene expression; scaffold attachment regions; origins of DNA replication; centromeres; and telomeres. Some regions appear to be mostly nonfunctional such as introns, pseudogenes, intergenic DNA, and fragments of transposons and viruses. These apparently non-functional regions take up most of the genome of many eukaryotes and many scientists think that they are junk DNA.

In genetics, a single-nucleotide polymorphism is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently large fraction of the population, many publications do not apply such a frequency threshold.
The International HapMap Project was an organization that aimed to develop a haplotype map (HapMap) of the human genome, to describe the common patterns of human genetic variation. HapMap is used to find genetic variants affecting health, disease and responses to drugs and environmental factors. The information produced by the project is made freely available for research.

Human genetic variation is the genetic differences in and among populations. There may be multiple variants of any given gene in the human population (alleles), a situation called polymorphism.
Zinc-finger nucleases (ZFNs) are artificial restriction enzymes generated by fusing a zinc finger DNA-binding domain to a DNA-cleavage domain. Zinc finger domains can be engineered to target specific desired DNA sequences and this enables zinc-finger nucleases to target unique sequences within complex genomes. By taking advantage of endogenous DNA repair machinery, these reagents can be used to precisely alter the genomes of higher organisms. Alongside CRISPR/Cas9 and TALEN, ZFN is a prominent tool in the field of genome editing.

In genomics, a genome-wide association study, also known as whole genome association study, is an observational study of a genome-wide set of genetic variants in different individuals to see if any variant is associated with a trait. GWA studies typically focus on associations between single-nucleotide polymorphisms (SNPs) and traits like major human diseases, but can equally be applied to any other genetic variants and any other organisms.
Gerald Mayer Rubin is an American biologist, notable for pioneering the use of transposable P elements in genetics, and for leading the public project to sequence the Drosophila melanogaster genome. Related to his genomics work, Rubin's lab is notable for development of genetic and genomics tools and studies of signal transduction and gene regulation. Rubin also serves as a vice president of the Howard Hughes Medical Institute and executive director of the Janelia Research Campus.

Whole genome sequencing (WGS), also known as full genome sequencing, complete genome sequencing, or entire genome sequencing, is the process of determining the entirety, or nearly the entirety, of the DNA sequence of an organism's genome at a single time. This entails sequencing all of an organism's chromosomal DNA as well as DNA contained in the mitochondria and, for plants, in the chloroplast.
WGAViewer is a bioinformatics software tool which is designed to visualize, annotate, and help interpret the results generated from a genome wide association study (GWAS). Alongside the P values of association, WGAViewer allows a researcher to visualize and consider other supporting evidence, such as the genomic context of the SNP, linkage disequilibrium (LD) with ungenotyped SNPs, gene expression database, and the evidence from other GWAS projects, when determining the potential importance of an individual SNP.
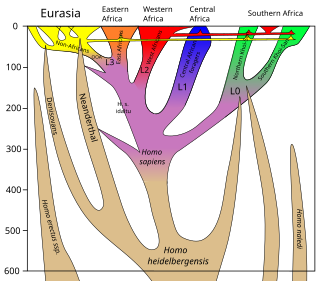
There is evidence for interbreeding between archaic and modern humans during the Middle Paleolithic and early Upper Paleolithic. The interbreeding happened in several independent events that included Neanderthals and Denisovans, as well as several unidentified hominins.

Interferon lambda 3 encodes the IFNL3 protein. IFNL3 was formerly named IL28B, but the Human Genome Organization Gene Nomenclature Committee renamed this gene in 2013 while assigning a name to the then newly discovered IFNL4 gene. Together with IFNL1 and IFNL2, these genes lie in a cluster on chromosomal region 19q13. IFNL3 shares ~96% amino-acid identity with IFNL2, ~80% identity with IFNL1 and ~30% identity with IFNL4.

David Matthew Altshuler is a clinical endocrinologist and human geneticist. He is Executive Vice President, Global Research and Chief Scientific Officer at Vertex Pharmaceuticals. Prior to joining Vertex in 2014, he was at the Broad Institute of Harvard and MIT, and was a Professor of Genetics and Medicine at Harvard Medical School, and in the Department of Biology at Massachusetts Institute of Technology. He was also a faculty member in the Department of Molecular Biology, Center for Human Genetic Research, and the Diabetes Unit, all at Massachusetts General Hospital. He was one of four Founding Core Members of the Broad Institute, and served as the Institute's Deputy Director, Chief Academic Officer, and Director of the Program in Medical and Population Genetics.
Single cell sequencing examines the sequence information from individual cells with optimized next-generation sequencing technologies, providing a higher resolution of cellular differences and a better understanding of the function of an individual cell in the context of its microenvironment. For example, in cancer, sequencing the DNA of individual cells can give information about mutations carried by small populations of cells. In development, sequencing the RNAs expressed by individual cells can give insight into the existence and behavior of different cell types. In microbial systems, a population of the same species can appear to be genetically clonal, but single-cell sequencing of RNA or epigenetic modifications can reveal cell-to-cell variability that may help populations rapidly adapt to survive in changing environments.

Wang Jun is a Chinese scientist, founder and CEO of iCarbonX, and former CEO of the Beijing Genomics Institute.
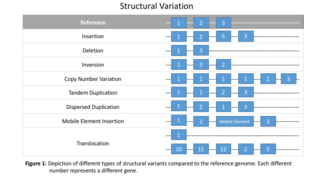
Structural variation in the human genome is operationally defined as genomic alterations, varying between individuals, that involve DNA segments larger than 1 kilo base (kb), and could be either microscopic or submicroscopic. This definition distinguishes them from smaller variants that are less than 1 kb in size such as short deletions, insertions, and single nucleotide variants.
In archaeogenetics, the term Ancient North Eurasian is the name given to a predominantly West-Eurasian ancestral component that represents descent from the people similar to the Mal'ta–Buret' culture and populations closely related to them, such as from Afontova Gora and the Yana Rhinoceros Horn Site.

Complex traits, also known as quantitative traits, are traits that do not behave according to simple Mendelian inheritance laws. More specifically, their inheritance cannot be explained by the genetic segregation of a single gene. Such traits show a continuous range of variation and are influenced by both environmental and genetic factors. Compared to strictly Mendelian traits, complex traits are far more common, and because they can be hugely polygenic, they are studied using statistical techniques such as QTL mapping rather than classical genetics methods. Examples of complex traits include height, circadian rhythms, enzyme kinetics, and many diseases including diabetes and Parkinson's disease. One major goal of genetic research today is to better understand the molecular mechanisms through which genetic variants act to influence complex traits.
In genetics, a haplotype block is a region of an organism's genome in which there is little evidence of a history of genetic recombination, and which contain only a small number of distinct haplotypes. According to the haplotype-block model, such blocks should show high levels of linkage disequilibrium and be separated from one another by numerous recombination events. The boundaries of haplotype blocks cannot be directly observed; they must instead be inferred indirectly through the use of algorithms. However, some evidence suggests that different algorithms for identifying haplotype blocks give very different results when used on the same data, though another study suggests that their results are generally consistent. The National Institutes of Health funded the HapMap project to catalog haplotype blocks throughout the human genome.












