
Microtubules are polymers of tubulin that form part of the cytoskeleton and provide structure and shape to eukaryotic cells. Microtubules can be as long as 50 micrometres, as wide as 23 to 27 nm and have an inner diameter between 11 and 15 nm. They are formed by the polymerization of a dimer of two globular proteins, alpha and beta tubulin into protofilaments that can then associate laterally to form a hollow tube, the microtubule. The most common form of a microtubule consists of 13 protofilaments in the tubular arrangement.
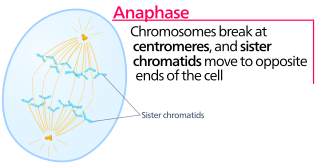
Anaphase, is the stage of mitosis after the process of metaphase, when replicated chromosomes are split and the newly-copied chromosomes are moved to opposite poles of the cell. Chromosomes also reach their overall maximum condensation in late anaphase, to help chromosome segregation and the re-formation of the nucleus.

In cell biology, the spindle apparatus refers to the cytoskeletal structure of eukaryotic cells that forms during cell division to separate sister chromatids between daughter cells. It is referred to as the mitotic spindle during mitosis, a process that produces genetically identical daughter cells, or the meiotic spindle during meiosis, a process that produces gametes with half the number of chromosomes of the parent cell.

Telophase is the final stage in both meiosis and mitosis in a eukaryotic cell. During telophase, the effects of prophase and prometaphase are reversed. As chromosomes reach the cell poles, a nuclear envelope is re-assembled around each set of chromatids, the nucleoli reappear, and chromosomes begin to decondense back into the expanded chromatin that is present during interphase. The mitotic spindle is disassembled and remaining spindle microtubules are depolymerized. Telophase accounts for approximately 2% of the cell cycle's duration.
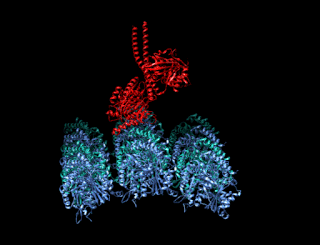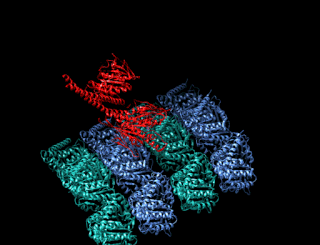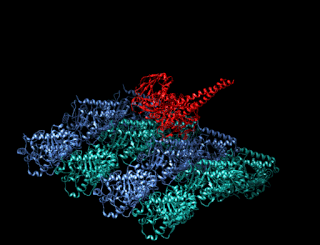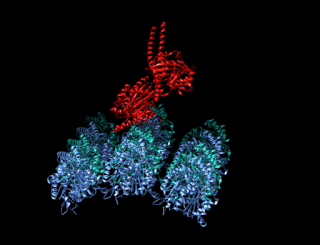
A kinesin is a protein belonging to a class of motor proteins found in eukaryotic cells.

The spindle checkpoint, also known as the metaphase-to-anaphase transition, the spindle assembly checkpoint (SAC), the metaphase checkpoint, or the mitotic checkpoint, is a cell cycle checkpoint during mitosis or meiosis that prevents the separation of the duplicated chromosomes (anaphase) until each chromosome is properly attached to the spindle. To achieve proper segregation, the two kinetochores on the sister chromatids must be attached to opposite spindle poles. Only this pattern of attachment will ensure that each daughter cell receives one copy of the chromosome. The defining biochemical feature of this checkpoint is the stimulation of the anaphase-promoting complex by M-phase cyclin-CDK complexes, which in turn causes the proteolytic destruction of cyclins and proteins that hold the sister chromatids together.

A kinetochore is a disc-shaped protein structure associated with duplicated chromatids in eukaryotic cells where the spindle fibers attach during cell division to pull sister chromatids apart. The kinetochore assembles on the centromere and links the chromosome to microtubule polymers from the mitotic spindle during mitosis and meiosis. The term kinetochore was first used in a footnote in a 1934 Cytology book by Lester W. Sharp and commonly accepted in 1936. Sharp's footnote reads: "The convenient term kinetochore has been suggested to the author by J. A. Moore", likely referring to John Alexander Moore who had joined Columbia University as a freshman in 1932.
A spindle poison, also known as a spindle toxin, is a poison that disrupts cell division by affecting the protein threads that connect the centromere regions of chromosomes, known as spindles. Spindle poisons effectively cease the production of new cells by interrupting the mitosis phase of cell division at the spindle assembly checkpoint (SAC). However, as numerous and varied as they are, spindle poisons are not yet 100% effective at ending the formation of tumors (neoplasms). Although not 100% effective, substantive therapeutic efficacy has been found in these types of chemotherapeutic treatments. The mitotic spindle is composed of microtubules that aid, along with regulatory proteins, each other in the activity of appropriately segregating replicated chromosomes. Certain compounds affecting the mitotic spindle have proven highly effective against solid tumors and hematological malignancies.
Mad2 is an essential spindle checkpoint protein. The spindle checkpoint system is a regulatory system that restrains progression through the metaphase-to-anaphase transition. The Mad2 gene was first identified in the yeast S. cerevisiae in a screen for genes which when mutated would confer sensitivity to microtubule poisons. The human orthologues of Mad2 were first cloned in a search for human cDNAs that would rescue the microtubule poison-sensitivity of a yeast strain in which a kinetochore binding protein was missing. The protein was shown to be present at unattached kinetochores and antibody inhibition studies demonstrated it was essential to execute a block in the metaphase-to-anaphase transition in response to the microtubule poison nocodazole. Subsequent cloning of the Xenopus laevis orthologue, facilitated by the sharing of the human sequence, allowed for the characterization of the mitotic checkpoint in egg extracts.

Aurora kinase B is a protein that functions in the attachment of the mitotic spindle to the centromere.
Polo-like kinases (Plks) are regulatory serine/threonin kinases of the cell cycle involved in mitotic entry, mitotic exit, spindle formation, cytokinesis, and meiosis. Only one Plk is found in the genomes of the fly Drosophila melanogaster (Polo), budding yeast (Cdc5) and fission yeast (Plo1). Vertebrates and other animals, however, have many Plk family members including Plk1, Plk2/Snk, Plk3/Prk/FnK, Plk4/Sak and Plk5. Of the vertebrate Plk family members, the mammalian Plk1 has been most extensively studied. During mitosis and cytokinesis, Plks associate with several structures including the centrosome, kinetochores, and the central spindle.

Kinesin-like protein KIF2C is a protein that in humans is encoded by the KIF2C gene.

Protein Regulator of cytokinesis 1 (PRC1) is a protein that in humans is encoded by the PRC1 gene and is involved in cytokinesis.

Kinesin-like protein KIFC1 is a protein that in humans is encoded by the KIFC1 gene.
The Kinesin 8 Family are a subfamily of the molecular motor proteins known as kinesins. Most kinesins transport materials or cargo around the cell while traversing along microtubule polymer tracks with the help of ATP-hydrolysis-created energy. The Kinesin 8 family has been shown to play an important role in chromosome alignment during mitosis. Kinesin 8 family members KIF18A in humans and Kip3 in yeast have been shown to be in vivo plus-end directed microtubule depolymerizers. During prometaphase of mitosis, the microtubules attach to the kinetochores of sister chromatids. Kinesin 8 is thought to play some role in this process, as knockdown of this protein via siRNA produces a phenotype of sister chromatids that are unable to align properly.

Mad1 is a non-essential protein which in yeast has a function in the spindle assembly checkpoint (SAC). This checkpoint monitors chromosome attachment to spindle microtubules and prevents cells from starting anaphase until the spindle is built up. The name Mad refers to the observation that mutant cells are mitotic arrest deficient (MAD) during microtubule depolymerization. Mad1 recruits the anaphase inhibitor Mad2 to unattached kinetochores and is essential for Mad2-Cdc20 complex formation in vivo but not in vitro. In vivo, Mad1 acts as a competitive inhibitor of the Mad2-Cdc20 complex. Mad1 is phosphorylated by Mps1 which then leads together with other activities to the formation of the mitotic checkpoint complex (MCC). Thereby it inhibits the activity of the anaphase-promoting complex/cyclosome (APC/C). Homologues of Mad1 are conserved in eukaryotes from yeast to mammals.
Biorientation is the phenomenon whereby microtubules emanating from different microtubule organizing centres (MTOCs) attach to kinetochores of sister chromatids. This results in the sister chromatids moving to opposite poles of the cell during cell division, and thus results in both daughter cells having the same genetic information.

Kinesin-like protein KIF1A, also known as axonal transporter of synaptic vesicles or microtubule-based motor KIF1A, is a protein that in humans is encoded by the KIF1A gene.

Kinesin-like protein KIF11 is a molecular motor protein that is essential in mitosis. In humans it is coded for by the gene KIF11. Kinesin-like protein KIF11 is a member of the kinesin superfamily, which are nanomotors that move along microtubule tracks in the cell. Named from studies in the early days of discovery, it is also known as Kinesin-5, or as BimC, Eg5 or N-2, based on the founding members of this kinesin family.
J. Richard McIntosh is a Distinguished Professor Emeritus in Molecular, Cellular, and Developmental Biology at the University of Colorado Boulder. McIntosh first graduated from Harvard with a BA in Physics in 1961, and again with a Ph.D. in Biophysics in 1968. He began his teaching career at Harvard but has spent most of his career at the University of Colorado Boulder. At the University of Colorado Boulder, McIntosh taught biology courses at both the undergraduate and graduate levels. Additionally, he created an undergraduate course in the biology of cancer towards the last several years of his teaching career. McIntosh's research career looks at a variety of things, including different parts of mitosis, microtubules, and motor proteins.












