Related Research Articles

Acidobacteriota is a phylum of Gram-negative bacteria. Its members are physiologically diverse and ubiquitous, especially in soils, but are under-represented in culture.

Sphingomonas was defined in 1990 as a group of Gram-negative, rod-shaped, chemoheterotrophic, strictly aerobic bacteria. They possess ubiquinone 10 as their major respiratory quinone, contain glycosphingolipids (GSLs), specifically ceramide, instead of lipopolysaccharide (LPS) in their cell envelopes, and typically produce yellow-pigmented colonies. The GSL serves to protect the bacteria from antibacterial substances. Unlike most Gram-negative bacteria, Sphingomonas cannot carry endotoxins due to the lack of lipopolysaccharides, and has a hydrophobic surface characterized by the short nature of the GSL's carbohydrate portion.

Sphingomonadaceae are a gram-negative bacterial family of the Alphaproteobacteria. An important feature is the presence of sphingolipids in the outer membrane of the cell wall. The cells are ovoid or rod-shaped. Others are also pleomorphic, i.e. the cells change the shape over time. Some species from Sphingomonadaceae family are dominant components of biofilms.

Filamentation is the anomalous growth of certain bacteria, such as Escherichia coli, in which cells continue to elongate but do not divide. The cells that result from elongation without division have multiple chromosomal copies.
Dehalococcoides is a genus of bacteria within class Dehalococcoidia that obtain energy via the oxidation of hydrogen and subsequent reductive dehalogenation of halogenated organic compounds in a mode of anaerobic respiration called organohalide respiration. They are well known for their great potential to remediate halogenated ethenes and aromatics. They are the only bacteria known to transform highly chlorinated dioxins, PCBs. In addition, they are the only known bacteria to transform tetrachloroethene to ethene.

Shewanella is the sole genus included in the marine bacteria family Shewanellaceae. Some species within it were formerly classed as Alteromonas. Shewanella consists of facultatively anaerobic Gram-negative rods, most of which are found in extreme aquatic habitats where the temperature is very low and the pressure is very high. Shewanella bacteria are a normal component of the surface flora of fish and are implicated in fish spoilage. Shewanella chilikensis, a species of the genus Shewanella commonly found in the marine sponges of Saint Martin's Island of the Bay of Bengal, Bangladesh.

16S ribosomal RNA is the RNA component of the 30S subunit of a prokaryotic ribosome. It binds to the Shine-Dalgarno sequence and provides most of the SSU structure.
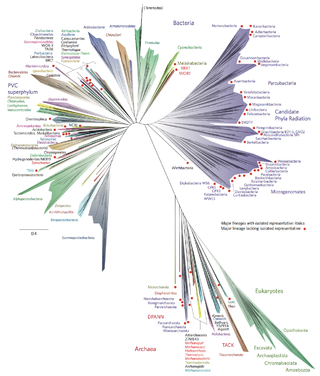
Bacterial phyla constitute the major lineages of the domain Bacteria. While the exact definition of a bacterial phylum is debated, a popular definition is that a bacterial phylum is a monophyletic lineage of bacteria whose 16S rRNA genes share a pairwise sequence identity of ~75% or less with those of the members of other bacterial phyla.
Saccharibacteria, formerly known as TM7, is a major bacterial lineage. It was discovered through 16S rRNA sequencing.

The class Zetaproteobacteria is the sixth and most recently described class of the Pseudomonadota. Zetaproteobacteria can also refer to the group of organisms assigned to this class. The Zetaproteobacteria were originally represented by a single described species, Mariprofundus ferrooxydans, which is an iron-oxidizing neutrophilic chemolithoautotroph originally isolated from Kamaʻehuakanaloa Seamount in 1996 (post-eruption). Molecular cloning techniques focusing on the small subunit ribosomal RNA gene have also been used to identify a more diverse majority of the Zetaproteobacteria that have as yet been unculturable.
"Candidatus Scalindua" is a bacterial genus, and a proposed member of the order Planctomycetales. These bacteria lack peptidoglycan in their cell wall and have a compartmentalized cytoplasm. They are ammonium oxidizing bacteria found in marine environments.
Geobacter sulfurreducens is a gram-negative metal- and sulphur-reducing proteobacterium. It is rod-shaped, aerotolerant anaerobe, non-fermentative, has flagellum and type four pili, and is closely related to Geobacter metallireducens. Geobacter sulfurreducens is an anaerobic species of bacteria that comes from the family of bacteria called Geobacteraceae. Under the genus of Geobacter, G. sulfurreducens is one out of twenty different species. The Geobacter genus was discovered by Derek R. Lovley in 1987. G. sulfurreducens was first isolated in Norman, Oklahoma, USA from materials found around the surface of a contaminated ditch.
Shewanella baltica DSS12 is a gram-negative bacterium. Its type strain is NCTC 10735. It is of particular importance in fish spoilage.
Sodalis is a genus of bacteria within the family Pectobacteriaceae. This genus contains several insect endosymbionts and also a free-living group. It is studied due to its potential use in the biological control of the tsetse fly. Sodalis is an important model for evolutionary biologists because of its nascent endosymbiosis with insects.

Bioluminescent bacteria are light-producing bacteria that are predominantly present in sea water, marine sediments, the surface of decomposing fish and in the gut of marine animals. While not as common, bacterial bioluminescence is also found in terrestrial and freshwater bacteria. These bacteria may be free living or in symbiosis with animals such as the Hawaiian Bobtail squid or terrestrial nematodes. The host organisms provide these bacteria a safe home and sufficient nutrition. In exchange, the hosts use the light produced by the bacteria for camouflage, prey and/or mate attraction. Bioluminescent bacteria have evolved symbiotic relationships with other organisms in which both participants benefit close to equally. Another possible reason bacteria use luminescence reaction is for quorum sensing, an ability to regulate gene expression in response to bacterial cell density.

James Ivor Prosser is a Professor in Environmental Microbiology in the Institute of Biological and Environmental Sciences at the University of Aberdeen.
Gabriele Berg is a biologist, biotechnologist and university lecturer in Environmental and Ecological Technology at the Technical University of Graz. Her research emphasis is on the development of sustainable methods of plant vitalisation with Bioeffectors and molecular analysis of microbial processes in the soil, particularly in the Rhizosphere.
Cytophagales is an order of non-spore forming, rod-shaped, Gram-negative bacteria that move through a gliding or flexing motion. These chemoorganotrophs are important remineralizers of organic materials into micronutrients. They are widely dispersed in the environment, found in ecosystems including soil, freshwater, seawater and sea ice. Cytophagales is included in the Bacteroidota phylum.

Plastic degradation in marine bacteria describes when certain pelagic bacteria break down polymers and use them as a primary source of carbon for energy. Polymers such as polyethylene (PE), polypropylene (PP), and polyethylene terephthalate (PET) are incredibly useful for their durability and relatively low cost of production, however it is their persistence and difficulty to be properly disposed of that is leading to pollution of the environment and disruption of natural processes. It is estimated that each year there are 9-14 million metric tons of plastic that are entering the ocean due to inefficient solutions for their disposal. The biochemical pathways that allow for certain microbes to break down these polymers into less harmful byproducts has been a topic of study to develop a suitable anti-pollutant.
Shimshon Belkin is an environmental microbiologist, a Professor Emeritus at the Department of Plant and Environmental Sciences at the Alexander Silberman Institute of Life Sciences of the Hebrew University of Jerusalem, Israel.
References
- 1 2 3 4 "Bacteria researcher receives annual award of DKK 5 million - DTU". www.dtu.dk. Retrieved 2022-01-15.
- 1 2 3 "Lone Gram Royal Academy". www.royalacademy.dk. March 31, 2021. Retrieved 2022-01-15.
- ↑ "Professor Lone Gram". www.royalacademy.dk. Retrieved 2022-01-15.
- ↑ Gram, Lone (1985). Konduktansmaling og mikrokalorimetri som hurtigmetoder i levnedsmiddelhygiejne: en undersgelse af fersk fisk (Thesis) (in Danish). Kobenhavn: Institut for Veterinr Mikrobiologi og Hygiejne, KVL. OCLC 476091721.
- ↑ Gram, Lone (1989). Identification, characterization, and inhibition of bacteria isolated from tropical fish (Thesis). Copenhagen, Denmark. OCLC 222426373.
- ↑ Gram, Lone; Trolle, Gunilla; Huss, Hans Henrik (1987-01-01). "Detection of specific spoilage bacteria from fish stored at low (0°C) and high (20°C) temperatures". International Journal of Food Microbiology. 4 (1): 65–72. doi:10.1016/0168-1605(87)90060-2. ISSN 0168-1605.
- ↑ Gram, L. (1993). "Inhibitory effect against pathogenic and spoilage bacteria of Pseudomonas strains isolated from spoiled and fresh fish". Applied and Environmental Microbiology. 59 (7): 2197–2203. Bibcode:1993ApEnM..59.2197G. doi:10.1128/aem.59.7.2197-2203.1993. PMC 182257 . PMID 8357253.
- ↑ Gram, Lone; Huss, Hans Henrik (1996-11-01). "Microbiological spoilage of fish and fish products". International Journal of Food Microbiology. Specific Spoilage Organisms. 33 (1): 121–137. doi:10.1016/0168-1605(96)01134-8. ISSN 0168-1605. PMID 8913813.
- ↑ Johansen, C.; Falholt, P.; Gram, L. (1997). "Enzymatic removal and disinfection of bacterial biofilms". Applied and Environmental Microbiology. 63 (9): 3724–3728. Bibcode:1997ApEnM..63.3724J. doi:10.1128/aem.63.9.3724-3728.1997. PMC 168680 . PMID 9293025.
- ↑ Maximilien, R; de Nys, R; Holmström, C; Gram, L; Givskov, M; Crass, K; Kjelleberg, S; Steinberg, Pd (1998). "Chemical mediation of bacterial surface colonisation by secondary metabolites from the red alga Delisea pulchra". Aquatic Microbial Ecology. 15: 233–246. doi: 10.3354/ame015233 . ISSN 0948-3055.
- ↑ Gram, Lone; Melchiorsen, Jette; Spanggaard, Bettina; Huber, Ingrid; Nielsen, Torben F. (1999-03-01). "Inhibition of Vibrio anguillarum byPseudomonas fluorescens AH2, a Possible Probiotic Treatment of Fish". Applied and Environmental Microbiology. 65 (3): 969–973. Bibcode:1999ApEnM..65..969G. doi:10.1128/AEM.65.3.969-973.1999. PMC 91130 . PMID 10049849.
- ↑ Gram, Lone; Christensen, Allan Beck; Ravn, Lars; Molin, Søren; Givskov, Michael (1999-08-01). "Production of Acylated Homoserine Lactones by Psychrotrophic Members of the Enterobacteriaceae Isolated from Foods". Applied and Environmental Microbiology. 65 (8): 3458–3463. Bibcode:1999ApEnM..65.3458G. doi:10.1128/AEM.65.8.3458-3463.1999. PMC 91519 . PMID 10427034. S2CID 30130863.
- ↑ Kingshott, Peter; Wei, Jiang; Bagge-Ravn, Dorthe; Gadegaard, Nikolaj; Gram, Lone (2003-08-01). "Covalent Attachment of Poly(ethylene glycol) to Surfaces, Critical for Reducing Bacterial Adhesion". Langmuir. 19 (17): 6912–6921. doi:10.1021/la034032m. ISSN 0743-7463.
- ↑ Vogel, Birte Fonnesbech; Venkateswaran, Kasthuri; Satomi, Masataka; Gram, Lone (2005). "Identification of Shewanella baltica as the Most Important H2S-Producing Species during Iced Storage of Danish Marine Fish". Applied and Environmental Microbiology. 71 (11): 6689–6697. Bibcode:2005ApEnM..71.6689V. doi:10.1128/AEM.71.11.6689-6697.2005. PMC 1287644 . PMID 16269698.
- ↑ Tachibana, Chris (2013-05-16). "Blue biotechnology wave - Nordic Life Science". Nordic Life Science – the leading Nordic life science news service. Retrieved 2022-01-15.
- ↑ Gram, Lone; Melchiorsen, Jette; Bruhn, Jesper Bartholin (2010). "Antibacterial Activity of Marine Culturable Bacteria Collected from a Global Sampling of Ocean Surface Waters and Surface Swabs of Marine Organisms". Marine Biotechnology. 12 (4): 439–451. doi:10.1007/s10126-009-9233-y. ISSN 1436-2228. PMID 19823914. S2CID 27071269.
- ↑ Sonnenschein, Eva C; Nielsen, Kristian F; D'Alvise, Paul; Porsby, Cisse H; Melchiorsen, Jette; Heilmann, Jens; Kalatzis, Panos G; López-Pérez, Mario; Bunk, Boyke; Spröer, Cathrin; Middelboe, Mathias (2017). "Global occurrence and heterogeneity of the Roseobacter-clade species Ruegeria mobilis". The ISME Journal. 11 (2): 569–583. doi:10.1038/ismej.2016.111. ISSN 1751-7362. PMC 5270555 . PMID 27552638.
- ↑ Giubergia, Sonia; Phippen, Christopher; Gotfredsen, Charlotte H.; Nielsen, Kristian Fog; Gram, Lone (2016-04-29). "Influence of Niche-Specific Nutrients on Secondary Metabolism in Vibrionaceae". Applied and Environmental Microbiology. 82 (13): 4035–4044. Bibcode:2016ApEnM..82.4035G. doi:10.1128/AEM.00730-16. PMC 4907173 . PMID 27129958.
- 1 2 "Lone Gram". Welcome to DTU Research Database. Retrieved 2022-01-15.
- ↑ "Professor Lone Gran decorated with the Order of Dannebrog - CeMiSt". Center for Microbial Secondary Metabolites. Retrieved 2022-01-15.
- ↑ Shekhar, Akarsh (2020-10-22). "Hans Christian Gram: 8 Facts About The Man Who Invented the Famous 'Stain Technique'". DailyHawker. Retrieved 2022-01-15.
- ↑ "Opening of the Danish National Research Foundation DNRF, Center of Excellence:Center for microbial secondary metabolites (Cemist)" (PDF). Danish National Research Foundation. February 27, 2017.