Related Research Articles

Antimicrobial resistance (AMR) occurs when microbes evolve mechanisms that protect them from the effects of antimicrobials. All classes of microbes can evolve resistance where the drugs are no longer effective. Fungi evolve antifungal resistance. Viruses evolve antiviral resistance. Protozoa evolve antiprotozoal resistance, and bacteria evolve antibiotic resistance. Together all of these come under the umbrella of antimicrobial resistance. Microbes resistant to multiple antimicrobials are called multidrug resistant (MDR) and are sometimes referred to as superbugs. Although antimicrobial resistance is a naturally occurring process, it is often the result of improper usage of the drugs and management of the infections.
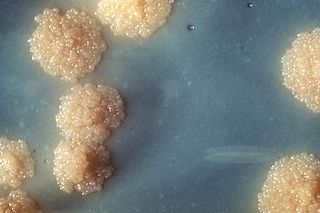
Mycobacterium tuberculosis, also known as Koch's bacillus, is a species of pathogenic bacteria in the family Mycobacteriaceae and the causative agent of tuberculosis. First discovered in 1882 by Robert Koch, M. tuberculosis has an unusual, waxy coating on its cell surface primarily due to the presence of mycolic acid. This coating makes the cells impervious to Gram staining, and as a result, M. tuberculosis can appear weakly Gram-positive. Acid-fast stains such as Ziehl–Neelsen, or fluorescent stains such as auramine are used instead to identify M. tuberculosis with a microscope. The physiology of M. tuberculosis is highly aerobic and requires high levels of oxygen. Primarily a pathogen of the mammalian respiratory system, it infects the lungs. The most frequently used diagnostic methods for tuberculosis are the tuberculin skin test, acid-fast stain, culture, and polymerase chain reaction.

Mycobacterium is a genus of over 190 species in the phylum Actinomycetota, assigned its own family, Mycobacteriaceae. This genus includes pathogens known to cause serious diseases in mammals, including tuberculosis and leprosy in humans. The Greek prefix myco- means 'fungus', alluding to this genus' mold-like colony surfaces. Since this genus has cell walls with a waxy lipid-rich outer layer that contains high concentrations of mycolic acid, acid-fast staining is used to emphasize their resistance to acids, compared to other cell types.

Rifampicin, also known as rifampin, is an ansamycin antibiotic used to treat several types of bacterial infections, including tuberculosis (TB), Mycobacterium avium complex, leprosy, and Legionnaires’ disease. It is almost always used together with other antibiotics with two notable exceptions: when given as a "preferred treatment that is strongly recommended" for latent TB infection; and when used as post-exposure prophylaxis to prevent Haemophilus influenzae type b and meningococcal disease in people who have been exposed to those bacteria. Before treating a person for a long period of time, measurements of liver enzymes and blood counts are recommended. Rifampicin may be given either by mouth or intravenously.
Multiple drug resistance (MDR), multidrug resistance or multiresistance is antimicrobial resistance shown by a species of microorganism to at least one antimicrobial drug in three or more antimicrobial categories. Antimicrobial categories are classifications of antimicrobial agents based on their mode of action and specific to target organisms. The MDR types most threatening to public health are MDR bacteria that resist multiple antibiotics; other types include MDR viruses, parasites.

Antimicrobial peptides (AMPs), also called host defence peptides (HDPs) are part of the innate immune response found among all classes of life. Fundamental differences exist between prokaryotic and eukaryotic cells that may represent targets for antimicrobial peptides. These peptides are potent, broad spectrum antimicrobials which demonstrate potential as novel therapeutic agents. Antimicrobial peptides have been demonstrated to kill Gram negative and Gram positive bacteria, enveloped viruses, fungi and even transformed or cancerous cells. Unlike the majority of conventional antibiotics it appears that antimicrobial peptides frequently destabilize biological membranes, can form transmembrane channels, and may also have the ability to enhance immunity by functioning as immunomodulators.

4-Aminosalicylic acid, also known as para-aminosalicylic acid (PAS) and sold under the brand name Paser among others, is an antibiotic primarily used to treat tuberculosis. Specifically it is used to treat active drug resistant tuberculosis together with other antituberculosis medications. It has also been used as a second line agent to sulfasalazine in people with inflammatory bowel disease such as ulcerative colitis and Crohn's disease. It is typically taken by mouth.

Rifabutin (Rfb) is an antibiotic used to treat tuberculosis and prevent and treat Mycobacterium avium complex. It is typically only used in those who cannot tolerate rifampin such as people with HIV/AIDS on antiretrovirals. For active tuberculosis it is used with other antimycobacterial medications. For latent tuberculosis it may be used by itself when the exposure was with drug-resistant TB.
The rpoB gene encodes the β subunit of bacterial RNA polymerase and the homologous plastid-encoded RNA polymerase (PEP). It codes for 1342 amino acids in E. coli, making it the second-largest polypeptide in the bacterial cell. It is targeted by the rifamycin family of antibacterials, such as rifampin. Mutations in rpoB that confer resistance to rifamycins do so by altering the protein's drug-binding residues, thereby reducing affinity for these antibiotics.

Ethionamide is an antibiotic used to treat tuberculosis. Specifically it is used, along with other antituberculosis medications, to treat active multidrug-resistant tuberculosis. It is no longer recommended for leprosy. It is taken by mouth.

A mycobacteriophage is a member of a group of bacteriophages known to have mycobacteria as host bacterial species. While originally isolated from the bacterial species Mycobacterium smegmatis and Mycobacterium tuberculosis, the causative agent of tuberculosis, more than 4,200 mycobacteriophage have since been isolated from various environmental and clinical sources. 2,042 have been completely sequenced. Mycobacteriophages have served as examples of viral lysogeny and of the divergent morphology and genetic arrangement characteristic of many phage types.
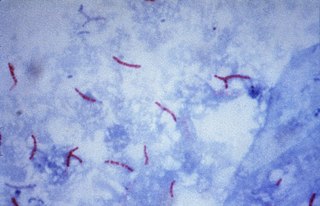
Multidrug-resistant tuberculosis (MDR-TB) is a form of tuberculosis (TB) infection caused by bacteria that are resistant to treatment with at least two of the most powerful first-line anti-TB medications (drugs): isoniazid and rifampin. Some forms of TB are also resistant to second-line medications, and are called extensively drug-resistant TB (XDR-TB).

Beta-lactamases are a family of enzymes involved in bacterial resistance to beta-lactam antibiotics. In bacterial resistance to beta-lactam antibiotics, the bacteria have beta-lactamase which degrade the beta-lactam rings, rendering the antibiotic ineffective. However, with beta-lactamase inhibitors, these enzymes on the bacteria are inhibited, thus allowing the antibiotic to take effect. Strategies for combating this form of resistance have included the development of new beta-lactam antibiotics that are more resistant to cleavage and the development of the class of enzyme inhibitors called beta-lactamase inhibitors. Although β-lactamase inhibitors have little antibiotic activity of their own, they prevent bacterial degradation of beta-lactam antibiotics and thus extend the range of bacteria the drugs are effective against.

Aminocoumarin is a class of antibiotics that act by an inhibition of the DNA gyrase enzyme involved in the cell division in bacteria. They are derived from Streptomyces species, whose best-known representative – Streptomyces coelicolor – was completely sequenced in 2002. The aminocoumarin antibiotics include:
The Xpert MTB/RIF is a cartridge-based nucleic acid amplification test (NAAT) for simultaneous rapid tuberculosis diagnosis and rapid antibiotic sensitivity test. It is an automated diagnostic test that can identify Mycobacterium tuberculosis (MTB) DNA and resistance to rifampicin (RIF). It was co-developed by the laboratory of Professor David Alland at the University of Medicine and Dentistry of New Jersey (UMDNJ), Cepheid Inc. and Foundation for Innovative New Diagnostics, with additional financial support from the US National Institutes of Health (NIH).
Computational Resources for Drug Discovery (CRDD) is one of the important silico modules of Open Source for Drug Discovery (OSDD). The CRDD web portal provides computer resources related to drug discovery on a single platform. It provides computational resources for researchers in computer-aided drug design, a discussion forum, and resources to maintain a wiki related to drug discovery, predict inhibitors, and predict the ADME-Tox property of molecules. One of the major objectives of CRDD is to promote open source software in the field of chemoinformatics and pharmacoinformatics.
Nagasuma Chandra is an Indian structural biologist, biochemist and a professor at the department of biochemistry of the Indian Institute of Science. She is known for her studies on Mycobacterium tuberculosis. The Department of Biotechnology of the Government of India awarded her the National Bioscience Award for Career Development, one of the highest Indian science awards, for her contributions to biosciences in 2008.
In molecular biology, MvirDB is a publicly available database that stores information on toxins, virulence factors and antibiotic resistance genes. Sources that this database uses for DNA and protein information include: Tox-Prot, SCORPION, the PRINTS Virulence Factors, VFDB, TVFac, Islander, ARGO and VIDA. The database provides a BLAST tool that allows the user to query their sequence against all DNA and protein sequences in MvirDB. Information on virulence factors can be obtained from the usage of the provided browser tool. Once the browser tool is used, the results are returned as a readable table that is organized by ascending E-Values, each of which are hyperlinked to their related page. MvirDB is implemented in an Oracle 10g relational database.
AMDD otherwise known as Antimicrobial Drug Database is a biological database that seeks to consolidate antibacterial and antifungal drug information from a variety of sources such as PubChem, PubChem Bioassay, ZINC, ChemDB and DrugBank in order to advance the field of treatment of resistance microbes. As of 2012, AMDD contains ~2900 antibacterial and ~1200 antifungal compounds. These compounds are organized via their description, target, format, bioassay, molecular weight, hydrogen bond donor, hydrogen bond acceptor and rotatable bond. AMDD was built on Apache server 2.2.11. The function of this database is to ultimately provide a comprehensive tool that is intuitively navigated such that development of novel antibacterial and antifungal compounds are facilitated.
References
- 1 2 3 4 Flandrois, Jean-Pierre; Lina, Gérard; Dumitrescu, Oana (December 2014). "MUBII-TB-DB: a database of mutations associated with antibiotic resistance in Mycobacterium tuberculosis". BMC Bioinformatics. 15 (1): 107. doi: 10.1186/1471-2105-15-107 . ISSN 1471-2105. PMC 4021062 . PMID 24731071.
- ↑ Heym, B (February 1997). "Multidrug resistance in Mycobacterium tuberculosis". International Journal of Antimicrobial Agents. 8 (1): 61–70. doi:10.1016/s0924-8579(96)00356-1. ISSN 0924-8579. PMID 18611785.
- ↑ Musser, J M (October 1995). "Antimicrobial agent resistance in mycobacteria: molecular genetic insights". Clinical Microbiology Reviews. 8 (4): 496–514. doi:10.1128/cmr.8.4.496. ISSN 0893-8512. PMC 172873 . PMID 8665467.
- ↑ Almeida Da Silva, P. E.; Palomino, J. C. (2011-07-01). "Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs". Journal of Antimicrobial Chemotherapy. 66 (7): 1417–1430. doi: 10.1093/jac/dkr173 . ISSN 0305-7453. PMID 21558086.
- ↑ Zhang, Y.; Yew, W-W. (2015-11-01). "Mechanisms of drug resistance in Mycobacterium tuberculosis: update 2015". The International Journal of Tuberculosis and Lung Disease. 19 (11): 1276–1289. doi:10.5588/ijtld.15.0389. ISSN 1027-3719. PMID 26467578.
- ↑ Sandgren, Andreas; Strong, Michael; Muthukrishnan, Preetika; Weiner, Brian K; Church, George M; Murray, Megan B (2009-02-10). "Tuberculosis Drug Resistance Mutation Database". PLOS Medicine. 6 (2): e1000002. doi: 10.1371/journal.pmed.1000002 . ISSN 1549-1676. PMC 2637921 . PMID 19209951.