
In cell biology, the nucleus is a membrane-bound organelle found in eukaryotic cells. Eukaryotes usually have a single nucleus, but a few cell types, such as mammalian red blood cells, have no nuclei, and a few others including osteoclasts have many. The main structures making up the nucleus are the nuclear envelope, a double membrane that encloses the entire organelle and isolates its contents from the cellular cytoplasm; and the nuclear matrix, a network within the nucleus that adds mechanical support, much like the cytoskeleton supports the cell as a whole.

Pyrrolysine is an α-amino acid that is used in the biosynthesis of proteins in some methanogenic archaea and bacteria; it is not present in humans. It contains an α-amino group, a carboxylic acid group. Its pyrroline side-chain is similar to that of lysine in being basic and positively charged at neutral pH.
An S-layer is a part of the cell envelope found in almost all archaea, as well as in many types of bacteria. It consists of a monomolecular layer composed of identical proteins or glycoproteins. This structure is built via self-assembly and encloses the whole cell surface. Thus, the S-layer protein can represent up to 15% of the whole protein content of a cell. S-layer proteins are poorly conserved or not conserved at all, and can differ markedly even between related species. Depending on species, the S-layers have a thickness between 5 and 25 nm and possess identical pores with 2–8 nm in diameter.

Porins are beta barrel proteins that cross a cellular membrane and act as a pore, through which molecules can diffuse. Unlike other membrane transport proteins, porins are large enough to allow passive diffusion, i.e., they act as channels that are specific to different types of molecules. They are present in the outer membrane of gram-negative bacteria and some gram-positive mycobacteria, the outer membrane of mitochondria, and the outer chloroplast membrane.

The Matrix-2 (M2) protein is a proton-selective viroporin, integral in the viral envelope of the influenza A virus. The channel itself is a homotetramer, where the units are helices stabilized by two disulfide bonds, and is activated by low pH. The M2 protein is encoded on the seventh RNA segment together with the M1 protein. Proton conductance by the M2 protein in influenza A is essential for viral replication.

Methanosarcina is a genus of euryarchaeote archaea that produce methane. These single-celled organisms are known as anaerobic methanogens that produce methane using all three metabolic pathways for methanogenesis. They live in diverse environments where they can remain safe from the effects of oxygen, whether on the earth's surface, in groundwater, in deep sea vents, and in animal digestive tracts. Methanosarcina grow in colonies.
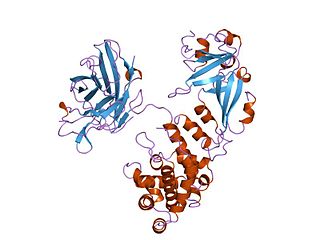
Diphtheria toxin is an exotoxin secreted by Corynebacterium, the pathogenic bacterium that causes diphtheria. The toxin gene is encoded by a prophage. The toxin causes the disease in humans by gaining entry into the cell cytoplasm and inhibiting protein synthesis.
Importin is a type of karyopherin that transports protein molecules from the cell's cytoplasm to the nucleus. It does so by binding to specific recognition sequences, called nuclear localization sequences (NLS).

The epithelial sodium channel (short: ENaC, also: amiloride-sensitive sodium channel) is a membrane-bound ion channel that is selectively permeable to the ions of sodium (Na+) and that is assembled as a heterotrimer composed of three homologous subunits α or δ, β, and γ, These subunits are encoded by four genes: SCNN1A, SCNN1B, SCNN1G, and SCNN1D. It is involved primarily in the reabsorption of sodium ions at the collecting ducts of the kidney's nephrons.

A DNA clamp, also known as a sliding clamp or β-clamp, is a protein complex that serves as a processivity-promoting factor in DNA replication. As a critical component of the DNA polymerase III holoenzyme, the clamp protein binds DNA polymerase and prevents this enzyme from dissociating from the template DNA strand. The clamp-polymerase protein–protein interactions are stronger and more specific than the direct interactions between the polymerase and the template DNA strand; because one of the rate-limiting steps in the DNA synthesis reaction is the association of the polymerase with the DNA template, the presence of the sliding clamp dramatically increases the number of nucleotides that the polymerase can add to the growing strand per association event. The presence of the DNA clamp can increase the rate of DNA synthesis up to 1,000-fold compared with a nonprocessive polymerase.

A colicin is a type of bacteriocin produced by and toxic to some strains of Escherichia coli. Colicins are released into the environment to reduce competition from other bacterial strains. Colicins bind to outer membrane receptors, using them to translocate to the cytoplasm or cytoplasmic membrane, where they exert their cytotoxic effect, including depolarisation of the cytoplasmic membrane, DNase activity, RNase activity, or inhibition of murein synthesis.

A protein domain is a conserved part of a given protein sequence and tertiary structure that can evolve, function, and exist independently of the rest of the protein chain. Each domain forms a compact three-dimensional structure and often can be independently stable and folded. Many proteins consist of several structural domains. One domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins.
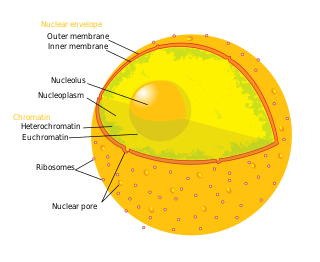
The nuclear envelope, also known as the nuclear membrane, is made up of two lipid bilayer membranes which in eukaryotic cells surrounds the nucleus, which encases the genetic material.
A domain of unknown function (DUF) is a protein domain that has no characterised function. These families have been collected together in the Pfam database using the prefix DUF followed by a number, with examples being DUF2992 and DUF1220. As of 2019, there are almost 4,000 DUF families within the Pfam database representing over 22% of known families. Some DUFs are not named using the nomenclature due to popular usage but are nevertheless DUFs.

In molecular biology, the CHAP domain is a region between 110 and 140 amino acids that is found in proteins from bacteria, bacteriophages, archaea and eukaryotes of the family Trypanosomidae. The domain is named after the acronym cysteine, histidine-dependent amidohydrolases/peptidases. Many of these proteins are uncharacterised, but it has been proposed that they may function mainly in peptidoglycan hydrolysis. The CHAP domain is found in a wide range of protein architectures; it is commonly associated with bacterial type SH3 domains and with several families of amidase domains. It has been suggested that CHAP domain containing proteins utilise a catalytic cysteine residue in a nucleophilic-attack mechanism.
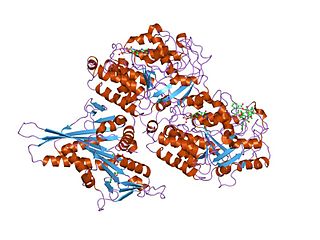
Tubulin/FtsZ family, GTPase domain is an evolutionary conserved protein domain.
An archaellum is a unique whip-like structure on the cell surface of many archaea. The name was proposed in 2012 following studies that showed it to be evolutionarily and structurally different from the bacterial and eukaryotic flagella. The archaellum is functionally the same – it can be rotated and is used to swim in liquid environments. The archaellum was found to be structurally similar to the type IV pilus.

In molecular biology, the YjeF N terminal is a protein domain found in the N-terminal of the protein, EDC3. The YjeF N-terminal domains occur either as single proteins or fusions with other domains and are commonly associated with enzymes. They help assemble the processing body (P-body) in preparation for mRNAdecay. Structural homology indicated it may have some similarity to the enzyme family, hydrolase.

Methanosarcina barkeri is the most fundamental species of the genus Methanosarcina, and their properties apply generally to the genus Methanosarcina. Methanosarcina barkeri can produce methane anaerobically through different metabolic pathways. M. barkeri can subsume a variety of molecules for ATP production, including methanol, acetate, methylamines, and different forms of hydrogen and carbon dioxide. Although it is a slow developer and is sensitive to change in environmental conditions, M. barkeri is able to grow in a variety of different substrates, adding to its appeal for genetic analysis. Additionally, M. barkeri is the first organism in which the amino acid pyrrolysine was found. Furthermore, two strains of M. barkeri, M. b. Fusaro and M. b. MS have been identified to possess an F-type ATPase along with an A-type ATPase.
Members of the H+, Na+-translocating Pyrophosphatase (M+-PPase) Family (TC# 3.A.10) are found in the vacuolar (tonoplast) membranes of higher plants, algae, and protozoa, and in both bacteria and archaea. They are therefore ancient enzymes.














