
In organic chemistry, a peptide bond is an amide type of covalent chemical bond linking two consecutive alpha-amino acids from C1 of one alpha-amino acid and N2 of another, along a peptide or protein chain.

Alamethicin is a channel-forming peptide antibiotic, produced by the fungus Trichoderma viride. It belongs to peptaibol peptides which contain the non-proteinogenic amino acid residue Aib. This residue strongly induces formation of alpha-helical structure. The peptide sequence is

Teicoplanin is an semisynthetic glycopeptide antibiotic with a spectrum of activity similar to vancomycin. Its mechanism of action is to inhibit bacterial cell wall peptidoglycan synthesis. It is used in the prophylaxis and treatment of serious infections caused by Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus and Enterococcus faecalis.
In organic chemistry, polyketides are a class of natural products derived from a precursor molecule consisting of a chain of alternating ketone and methylene groups: [−C(=O)−CH2−]n. First studied in the early 20th century, discovery, biosynthesis, and application of polyketides has evolved. It is a large and diverse group of secondary metabolites caused by its complex biosynthesis which resembles that of fatty acid synthesis. Because of this diversity, polyketides can have various medicinal, agricultural, and industrial applications. Many polyketides are medicinal or exhibit acute toxicity. Biotechnology has enabled discovery of more naturally-occurring polyketides and evolution of new polyketides with novel or improved bioactivity.
Nonribosomal peptides (NRP) are a class of peptide secondary metabolites, usually produced by microorganisms like bacteria and fungi. Nonribosomal peptides are also found in higher organisms, such as nudibranchs, but are thought to be made by bacteria inside these organisms. While there exist a wide range of peptides that are not synthesized by ribosomes, the term nonribosomal peptide typically refers to a very specific set of these as discussed in this article.
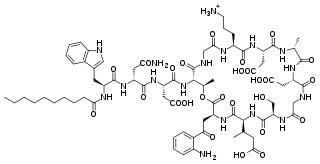
Daptomycin, sold under the brand name Cubicin among others, is a lipopeptide antibiotic used in the treatment of systemic and life-threatening infections caused by Gram-positive organisms.

Viomycin is a member of the tuberactinomycin family, a group of nonribosomal peptide antibiotics exhibiting anti-tuberculosis activity. The tuberactinomycin family is an essential component in the drug cocktail currently used to fight infections of Mycobacterium tuberculosis. Viomycin was the first member of the tuberactinomycins to be isolated and identified, and was used to treat TB until it was replaced by the less toxic, but structurally related compound, capreomycin. The tuberactinomycins target bacterial ribosomes, binding RNA and disrupting bacterial protein synthesis and certain forms of RNA splicing. Viomycin is produced by the actinomycete Streptomyces puniceus.

Gramicidin S or Gramicidin Soviet is an antibiotic that is effective against some gram-positive and gram-negative bacteria as well as some fungi.
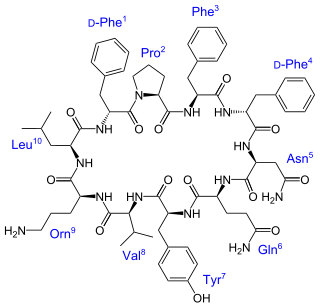
Tyrocidine is a mixture of cyclic decapeptides produced by the bacteria Brevibacillus brevis found in soil. It can be composed of 4 different amino acid sequences, giving tyrocidine A–D. Tyrocidine is the major constituent of tyrothricin, which also contains gramicidin. Tyrocidine was the first commercially available antibiotic, but has been found to be toxic toward human blood and reproductive cells. The function of tyrocidine within its host B. brevis is thought to be regulation of sporulation.
In enzymology, a 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase (EC 1.3.1.28) is an enzyme that catalyzes the chemical reaction
2,3-Dihydroxybenzoic acid is a natural phenol found in Phyllanthus acidus and in the aquatic fern Salvinia molesta. It is also abundant in the fruits of Flacourtia inermis. It is a dihydroxybenzoic acid, a type of organic compound.
Zwittermicin A is an antibiotic that has been identified from the bacterium Bacillus cereus UW85. It is a molecule of interest to agricultural industry because it has the potential to suppress plant disease due to its broad spectrum activity against certain gram positive and gram negative prokaryotic micro-organisms. The molecule is also of interest from a metabolic perspective because it represents a new structural class of antibiotic and suggests a crossover between polyketide and non-ribosomal peptide biosynthetic pathways. Zwittermicin A is linear aminopolyol.
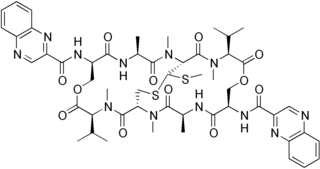
Echinomycin is a peptide antibiotic. It is a dimer of two peptides creating a cyclic structure. It contains a bicyclic aromatic chromophore that is attached to the dimerized cyclic peptide core and a thioacetal bridge. It intercalates into DNA at two specific sites, thereby blocking the binding of hypoxia inducible factor 1 alpha (HIF1alpha).

Hectochlorin is a lipopeptide that exhibits potent antifungal activity against C. albicans and a number of plants pathogens, as well as inhibiting growth of human cell lines by hyperpolymerization of actin. It was originally isolated from the filamentous cyanobacterium Moorea producens JHB, collected from Hector Bay, Jamaica, 1996, which is a strain also known for being the producer of other two potent biomolecules named Jamaicamide A and Cryptomaldamide. Due to its activity against plants pathogens, synthetic efforts elucidated the compound’s total synthesis in 2002. Moorea species are normally the main component of the dietary of some sea hares, which concentrate the cyanobacterial metabolites as a mechanism of defense from predators. Therefore, in 2005, hectochlorin was re-isolated from the Thai sea hare Bursatella leachii, along with a new analogue, deacetylhectochlorin. Another reisolation of hectochlorin was reported in 2013, from another Moorea producens strain (RS05), isolated from the Red Sea, surprising in a non-tropical environment, as opposed to the other Moorea strains isolated before. The predicted biosynthesis of hectochlorin was published in 2007 and consists in a hybrid NRPS-PKS, with a hexanoic acid as start unit that becomes halogenated twice in the position 5, producing fairly rare gem-dichloro group, that along with two 2,3-dihydroxyisovaleric acid (DHIV) units compose a very interesting bioactive molecule.

Bacillibactin is a catechol-based siderophore secreted by members of the genus Bacillus, including Bacillus anthracis and Bacillus subtilis. It is involved in the chelation of ferric iron (Fe3+) from the surrounding environment and is subsequently transferred into the bacterial cytoplasm via the use of ABC transporters.
Ribosomally synthesized and post-translationally modified peptides (RiPPs), also known as ribosomal natural products, are a diverse class of natural products of ribosomal origin. Consisting of more than 20 sub-classes, RiPPs are produced by a variety of organisms, including prokaryotes, eukaryotes, and archaea, and they possess a wide range of biological functions.

Vibriobactin is a catechol siderophore that helps the microbial system to acquire iron. It was first isolated from Vibrio cholerae.
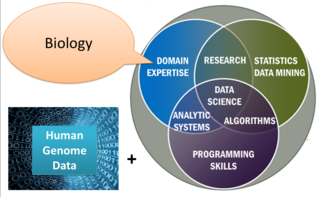
Genome mining describes the exploitation of genomic information for the discovery of biosynthetic pathways of natural products and their possible interactions. It depends on computational technology and bioinformatics tools. The mining process relies on a huge amount of data accessible in genomic databases. By applying data mining algorithms, the data can be used to generate new knowledge in several areas of medicinal chemistry, such as discovering novel natural products.

Tilivalline is a nonribosomal enterotoxin and was the first naturally occurring pyrrolobenzodiazepine (PBD) to be associated with disease in the human intestine. Previous work has shown that PBD tilivalline produced by Klebsiella oxytoca was linked to the pathogenesis of colitis in animal model of antibiotic-associated hemorrhagic colitis (AAHC). Since the enteric bacterium K. oxytoca is part of the intestinal microbiota and tilivalline causes the pathogenesis of colitis, it is important to understand the biosynthesis and regulation of tilivalline activity.

Petrobactin is a bis-catechol siderophore found in M. hydrocarbonoclasticus, A. macleodii, and the anthrax-producing B. anthracis. Like other siderophores petrobactin is a highly specific iron(III) transport ligand, contributing to the marine microbial uptake of environmental iron.















