Related Research Articles
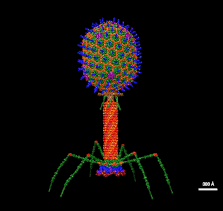
A bacteriophage, also known informally as a phage, is a virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν, meaning "to devour". Bacteriophages are composed of proteins that encapsulate a DNA or RNA genome, and may have structures that are either simple or elaborate. Their genomes may encode as few as four genes and as many as hundreds of genes. Phages replicate within the bacterium following the injection of their genome into its cytoplasm.
Enterobacteria phage λ is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell.

A prophage is a bacteriophage genome inserted and integrated into the circular bacterial DNA chromosome or exists as an extrachromosomal plasmid. This is a latent form of a phage, in which the viral genes are present in the bacterium without causing disruption of the bacterial cell. Pro means ''before'', so, prophage means the stage of a virus in the form of genome inserted into host DNA before being activated inside the host.

Phage therapy, viral phage therapy, or phagotherapy is the therapeutic use of bacteriophages to treat pathogenic bacterial infections. Bacteriophages, known as phages, are a form of viruses. Phages attach to bacterial cells, and inject a viral genome into the cell. The viral genome effectively replaces the bacterial genome, halting the bacterial infection. The bacterial cell causing the infection is unable to reproduce, and instead produces additional phages. Phages are very selective in the strains of bacteria they are effective against. Advantages include reduced side-effects and reduced risk of the bacterium's developing resistance. Disadvantages include the difficulty of finding an effective phage for a particular infection. However, virulent phages can be isolated much more quickly than other compounds and natural products due to the fact that they can be isolated from the environment with ease. In addition to this, development of standardized manufacturing processes would make lab to clinic delivery of phages much quicker.

Transduction is the process by which foreign DNA is introduced into a cell by a virus or viral vector. An example is the viral transfer of DNA from one bacterium to another and hence an example of horizontal gene transfer. Transduction does not require physical contact between the cell donating the DNA and the cell receiving the DNA, and it is DNase resistant. Transduction is a common tool used by molecular biologists to stably introduce a foreign gene into a host cell's genome.

The lytic cycle is one of the two cycles of viral reproduction, the other being the lysogenic cycle. The lytic cycle results in the destruction of the infected cell and its membrane. Bacteriophages that only use the lytic cycle are called virulent phages.

Escherichia virus T4 is a species of bacteriophages that infect Escherichia coli bacteria. It is a double-stranded DNA virus in the subfamily Tevenvirinae from the family Myoviridae. T4 is capable of undergoing only a lytic lifecycle and not the lysogenic lifecycle. The species was formerly named T-even bacteriophage, a name which also encompasses, among other strains, Enterobacteria phage T2, Enterobacteria phage T4 and Enterobacteria phage T6.

Filamentous bacteriophage is a family of viruses (Inoviridae) that infect bacteria. The phages are named for their filamentous shape, a worm-like chain, about 6 nm in diameter and about 1000-2000 nm long. The coat of the virion comprises five types of viral protein, which are located during phage assembly in the inner membrane of the host bacteria, and are added to the nascent virion as it extrudes through the membrane. The simplicity of this family makes it an attractive model system to study fundamental aspects of molecular biology, and it has also proven useful as a tool in immunology and nanotechnology.

Caudovirales is an order of viruses known as the tailed bacteriophages. Under the Baltimore classification scheme, the Caudovirales are group I viruses as they have double stranded DNA (dsDNA) genomes, which can be anywhere from 18,000 base pairs to 500,000 base pairs in length. The virus particles have a distinct shape; each virion has an icosahedral head that contains the viral genome, and is attached to a flexible tail by a connector protein. The order encompasses a wide range of viruses, many containing genes of similar nucleotide sequence and function. However, some tailed bacteriophage genomes can vary quite significantly in nucleotide sequence, even among the same genus. Due to their characteristic structure and possession of potentially homologous genes, it is believed these bacteriophages possess a common origin.
In virology, temperate refers to the ability of some bacteriophages to display a lysogenic life cycle. Many temperate phages can integrate their genomes into their host bacterium's chromosome, together becoming a lysogen as the phage genome becomes a prophage. A temperate phage is also able to undergo a productive, typically lytic life cycle, where the prophage is expressed, replicates the phage genome, and produces phage progeny, which then leave the bacterium. With phage the term virulent is often used as an antonym to temperate, but more strictly a virulent phage is one that has lost its ability to display lysogeny through mutation rather than a phage lineage with no genetic potential to ever display lysogeny.

Lysogeny, or the lysogenic cycle, is one of two cycles of viral reproduction. Lysogeny is characterized by integration of the bacteriophage nucleic acid into the host bacterium's genome or formation of a circular replicon in the bacterial cytoplasm. In this condition the bacterium continues to live and reproduce normally. The genetic material of the bacteriophage, called a prophage, can be transmitted to daughter cells at each subsequent cell division, and at later events can release it, causing proliferation of new phages via the lytic cycle. Lysogenic cycles can also occur in eukaryotes, although the method of DNA incorporation is not fully understood.
Salmonella virus P22 is a bacteriophage in the Podoviridae family that infects Salmonella typhimurium. Like many phages, it has been used in molecular biology to induce mutations in cultured bacteria and to introduce foreign genetic material. P22 has been used in generalized transduction and is an important tool for investigating Salmonella genetics.
P1 is a temperate bacteriophage that infects Escherichia coli and some other bacteria. When undergoing a lysogenic cycle the phage genome exists as a plasmid in the bacterium unlike other phages that integrate into the host DNA. P1 has an icosahedral head containing the DNA attached to a contractile tail with six tail fibers. The P1 phage has gained research interest because it can be used to transfer DNA from one bacterial cell to another in a process known as transduction. As it replicates during its lytic cycle it captures fragments of the host chromosome. If the resulting viral particles are used to infect a different host the captured DNA fragments can be integrated into the new host's genome. This method of in vivo genetic engineering was widely used for many years and is still used today, though to a lesser extent. P1 can also be used to create the P1-derived artificial chromosome cloning vector which can carry relatively large fragments of DNA. P1 encodes a site-specific recombinase, Cre, that is widely used to carry out cell-specific or time-specific DNA recombination by flanking the target DNA with loxP sites.
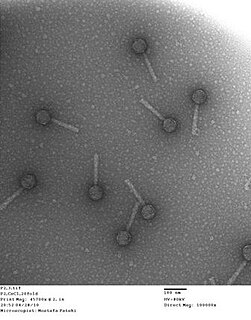
Bacteriophage P2, scientific name Escherichia virus P2, is a temperate phage that infects E. coli. It is a tailed virus with a contractile sheath and is thus classified in the genus Peduovirus, subfamily Peduovirinae, family Myoviridae within order Caudovirales. This genus of viruses includes many P2-like phages as well as the satellite phage P4.
The CTXφ bacteriophage is a filamentous bacteriophage. It is a positive-strand DNA virus with single-stranded DNA (ssDNA).
Bacteriophage T12 is a bacteriophage that infects the bacterial species Streptococcus pyogenes. It is a proosed species of the family Siphoviridae in order Caudovirales . It converts a harmless strain of bacteria into a virulent strain. It carries the speA gene which codes for erythrogenic toxin A. speA is also known as streptococcal pyogenic exotoxin A, scarlet fever toxin A, or even scarlatinal toxin. Note that the name of the gene "speA" is italicized; the name of the toxin "speA" is not italicized. Erythrogenic toxin A converts a harmless, non-virulent strain of Streptococcus pyogenes to a virulent strain through lysogeny, a life cycle which is characterized by the ability of the genome to become a part of the host cell and be stably maintained there for generations. Phages with a lysogenic life cycle are also called temperate phages. Bacteriophage T12, proposed member of family Siphoviridae including related speA-carrying bacteriophages, is also a prototypic phage for all the speA-carrying phages of Streptococcus pyogenes, meaning that its genome is the prototype for the genomes of all such phages of S. pyogenes. It is the main suspect as the cause of scarlet fever, an infectious disease that affects small children.
The Ff phages are filamentous phages that infect Gram-negative bacteria bearing the F episome. The term is used to refer to the closely related group of phages including the f1, fd and M13 phages. The If1 and Ike phage are also closely related to this group.

cII or transcriptional activator II is a DNA-binding protein and important transcription factor in the life cycle of lambda phage. It is encoded in the lambda phage genome by the 291 base pair cII gene. cII plays a key role in determining whether the bacteriophage will incorporate its genome into its host and lie dormant (lysogeny), or replicate and kill the host (lysis).
The Pseudomonas phage F116 holin is a non-characterized holin homologous to one in Neisseria gonorrheae that has been characterized. This protein is the prototype of the Pseudomonasphage F116 holin family, which is a member of the Holin Superfamily II. Bioinformatic analysis of the genome sequence of N. gonorrhoeae revealed the presence of nine probable prophage islands. The genomic sequence of FA1090 identified five genomic regions that are related to dsDNA lysogenic phage. The DNA sequences from NgoPhi1, NgoPhi2 and NgoPhi3 contained regions of identity. A region of NgoPhi2 showed high similarity with the Pseudomonas aeruginosa generalized transducing phage F116. NgoPhi1 and NgoPhi2 encode functionally active phages. The holin gene of NgoPhi1, when expressed in E. coli, could substitute for the phage lambda S gene.
Arbitrium is a viral peptide produced by bacteriophages to communicate with each other. It is six amino acids long, and is produced when a phage infects a bacterial host. It signals to other phages that a host has been infected.
References
- ↑ Brüssow, Harald; Canchaya, Carlos; Hardt, Wolf-Dietrich (September 2004). "Phages and the Evolution of Bacterial Pathogens: from Genomic Rearrangements to Lysogenic Conversion". Microbiol Mol Biol Rev. 68 (3): 560–602. doi:10.1128/mmbr.68.3.560-602.2004. PMC 515249 .
- ↑ Cumby, N; Davidson, AR; Maxwell, KL (2012). "The moron comes of age". Bacteriophage. 2 (4): 225–228. doi:10.4161/bact.23146. PMC 3594210 . PMID 23739268.
| This virus-related article is a stub. You can help Wikipedia by expanding it. |