
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria; however, plasmids are sometimes present in archaea and eukaryotic organisms. In nature, plasmids often carry genes that benefit the survival of the organism and confer selective advantage such as antibiotic resistance. While chromosomes are large and contain all the essential genetic information for living under normal conditions, plasmids are usually very small and contain only additional genes that may be useful in certain situations or conditions. Artificial plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms. In the laboratory, plasmids may be introduced into a cell via transformation. Synthetic plasmids are available for procurement over the internet.

A cloning vector is a small piece of DNA that can be stably maintained in an organism, and into which a foreign DNA fragment can be inserted for cloning purposes. The cloning vector may be DNA taken from a virus, the cell of a higher organism, or it may be the plasmid of a bacterium. The vector contains features that allow for the convenient insertion of a DNA fragment into the vector or its removal from the vector, for example through the presence of restriction sites. The vector and the foreign DNA may be treated with a restriction enzyme that cuts the DNA, and DNA fragments thus generated contain either blunt ends or overhangs known as sticky ends, and vector DNA and foreign DNA with compatible ends can then be joined together by molecular ligation. After a DNA fragment has been cloned into a cloning vector, it may be further subcloned into another vector designed for more specific use.
Site-directed mutagenesis is a molecular biology method that is used to make specific and intentional mutating changes to the DNA sequence of a gene and any gene products. Also called site-specific mutagenesis or oligonucleotide-directed mutagenesis, it is used for investigating the structure and biological activity of DNA, RNA, and protein molecules, and for protein engineering.

In molecular biology, a library is a collection of DNA fragments that is stored and propagated in a population of micro-organisms through the process of molecular cloning. There are different types of DNA libraries, including cDNA libraries, genomic libraries and randomized mutant libraries. DNA library technology is a mainstay of current molecular biology, genetic engineering, and protein engineering, and the applications of these libraries depend on the source of the original DNA fragments. There are differences in the cloning vectors and techniques used in library preparation, but in general each DNA fragment is uniquely inserted into a cloning vector and the pool of recombinant DNA molecules is then transferred into a population of bacteria or yeast such that each organism contains on average one construct. As the population of organisms is grown in culture, the DNA molecules contained within them are copied and propagated.
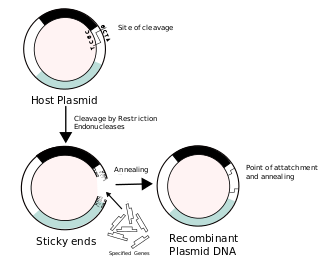
Recombinant DNA (rDNA) molecules are DNA molecules formed by laboratory methods of genetic recombination that bring together genetic material from multiple sources, creating sequences that would not otherwise be found in the genome.
A DNA construct is an artificially-designed segment of DNA borne on a vector that can be used to incorporate genetic material into a target tissue or cell. A DNA construct contains a DNA insert, called a transgene, delivered via a transformation vector which allows the insert sequence to be replicated and/or expressed in the target cell. This gene can be cloned from a naturally occurring gene, or synthetically constructed. The vector can be delivered using physical, chemical or viral methods. Typically, the vectors used in DNA constructs contain an origin of replication, a multiple cloning site, and a selectable marker. Certain vectors can carry additional regulatory elements based on the expression system involved.
A restriction digest is a procedure used in molecular biology to prepare DNA for analysis or other processing. It is sometimes termed DNA fragmentation. Hartl and Jones describe it this way:
This enzymatic technique can be used for cleaving DNA molecules at specific sites, ensuring that all DNA fragments that contain a particular sequence at a particular location have the same size; furthermore, each fragment that contains the desired sequence has the sequence located at exactly the same position within the fragment. The cleavage method makes use of an important class of DNA-cleaving enzymes isolated primarily from bacteria. These enzymes are called restriction endonucleases or restriction enzymes, and they are able to cleave DNA molecules at the positions at which particular short sequences of bases are present.

In molecular biology, subcloning is a technique used to move a particular DNA sequence from a parent vector to a destination vector.
A genomic library is a collection of the total genomic DNA from a single organism. The DNA is stored in a population of identical vectors, each containing a different insert of DNA. In order to construct a genomic library, the organism's DNA is extracted from cells and then digested with a restriction enzyme to cut the DNA into fragments of a specific size. The fragments are then inserted into the vector using DNA ligase. Next, the vector DNA can be taken up by a host organism - commonly a population of Escherichia coli or yeast - with each cell containing only one vector molecule. Using a host cell to carry the vector allows for easy amplification and retrieval of specific clones from the library for analysis.
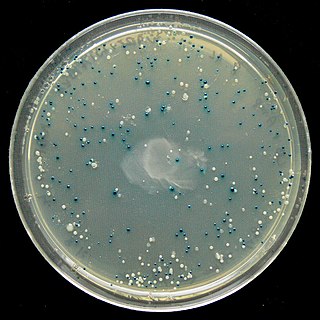
The blue–white screen is a screening technique that allows for the rapid and convenient detection of recombinant bacteria in vector-based molecular cloning experiments. This method of screening is usually performed using a suitable bacterial strain, but other organisms such as yeast may also be used. DNA of transformation is ligated into a vector. The vector is then inserted into a competent host cell viable for transformation, which are then grown in the presence of X-gal. Cells transformed with vectors containing recombinant DNA will produce white colonies; cells transformed with non-recombinant plasmids grow into blue colonies.

Artificial gene synthesis, or simply gene synthesis, refers to a group of methods that are used in synthetic biology to construct and assemble genes from nucleotides de novo. Unlike DNA synthesis in living cells, artificial gene synthesis does not require template DNA, allowing virtually any DNA sequence to be synthesized in the laboratory. It comprises two main steps, the first of which is solid-phase DNA synthesis, sometimes known as DNA printing. This produces oligonucleotide fragments that are generally under 200 base pairs. The second step then involves connecting these oligonucleotide fragments using various DNA assembly methods. Because artificial gene synthesis does not require template DNA, it is theoretically possible to make a completely synthetic DNA molecule with no limits on the nucleotide sequence or size.

Functional cloning is a molecular cloning technique that relies on prior knowledge of the encoded protein’s sequence or function for gene identification. In this assay, a genomic or cDNA library is screened to identify the genetic sequence of a protein of interest. Expression cDNA libraries may be screened with antibodies specific for the protein of interest or may rely on selection via the protein function. Historically, the amino acid sequence of a protein was used to prepare degenerate oligonucleotides which were then probed against the library to identify the gene encoding the protein of interest. Once candidate clones carrying the gene of interest are identified, they are sequenced and their identity is confirmed. This method of cloning allows researchers to screen entire genomes without prior knowledge of the location of the gene or the genetic sequence.
In molecular cloning, a vector is any particle used as a vehicle to artificially carry a foreign nucleic sequence – usually DNA – into another cell, where it can be replicated and/or expressed. A vector containing foreign DNA is termed recombinant DNA. The four major types of vectors are plasmids, viral vectors, cosmids, and artificial chromosomes. Of these, the most commonly used vectors are plasmids. Common to all engineered vectors have an origin of replication, a multicloning site, and a selectable marker.
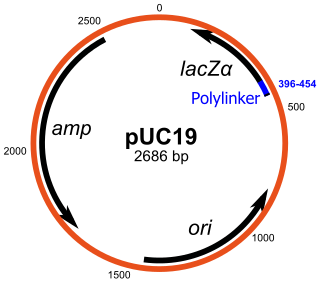
pUC19 is one of a series of plasmid cloning vectors created by Joachim Messing and co-workers. The designation "pUC" is derived from the classical "p" prefix and the abbreviation for the University of California, where early work on the plasmid series had been conducted. It is a circular double stranded DNA and has 2686 base pairs. pUC19 is one of the most widely used vector molecules as the recombinants, or the cells into which foreign DNA has been introduced, can be easily distinguished from the non-recombinants based on color differences of colonies on growth media. pUC18 is similar to pUC19, but the MCS region is reversed.
Transposons are semi-parasitic DNA sequences which can replicate and spread through the host's genome. They can be harnessed as a genetic tool for analysis of gene and protein function. The use of transposons is well-developed in Drosophila and in Thale cress and bacteria such as Escherichia coli.

Molecular cloning is a set of experimental methods in molecular biology that are used to assemble recombinant DNA molecules and to direct their replication within host organisms. The use of the word cloning refers to the fact that the method involves the replication of one molecule to produce a population of cells with identical DNA molecules. Molecular cloning generally uses DNA sequences from two different organisms: the species that is the source of the DNA to be cloned, and the species that will serve as the living host for replication of the recombinant DNA. Molecular cloning methods are central to many contemporary areas of modern biology and medicine.

Genetic engineering techniques allow the modification of animal and plant genomes. Techniques have been devised to insert, delete, and modify DNA at multiple levels, ranging from a specific base pair in a specific gene to entire genes. There are a number of steps that are followed before a genetically modified organism (GMO) is created. Genetic engineers must first choose what gene they wish to insert, modify, or delete. The gene must then be isolated and incorporated, along with other genetic elements, into a suitable vector. This vector is then used to insert the gene into the host genome, creating a transgenic or edited organism.
Addgene is a non-profit plasmid repository. Addgene facilitates the exchange of genetic material between laboratories by offering plasmids and their associated cloning data to not-for-profit laboratories around the world. Addgene provides a free online database of plasmid cloning information and references, including lists of commonly used vector backbones, popular lentiviral plasmids and molecular cloning protocols.

In molecular biology, ligation is the joining of two nucleic acid fragments through the action of an enzyme. It is an essential laboratory procedure in the molecular cloning of DNA, whereby DNA fragments are joined to create recombinant DNA molecules (such as when a foreign DNA fragment is inserted into a plasmid). The ends of DNA fragments are joined by the formation of phosphodiester bonds between the 3'-hydroxyl of one DNA terminus with the 5'-phosphoryl of another. RNA may also be ligated similarly. A co-factor is generally involved in the reaction, and this is usually ATP or NAD+.

Golden Gate Cloning or Golden Gate assembly is a molecular cloning method that allows a researcher to simultaneously and directionally assemble multiple DNA fragments into a single piece using Type IIS restriction enzymes and T4 DNA ligase. This assembly is performed in vitro. Most commonly used Type IIS enzymes include BsaI, BsmBI, and BbsI.














