
Antiviral drugs are a class of medication used for treating viral infections. Most antivirals target specific viruses, while a broad-spectrum antiviral is effective against a wide range of viruses. Antiviral drugs are a class of antimicrobials, a larger group which also includes antibiotic, antifungal and antiparasitic drugs, or antiviral drugs based on monoclonal antibodies. Most antivirals are considered relatively harmless to the host, and therefore can be used to treat infections. They should be distinguished from virucides, which are not medication but deactivate or destroy virus particles, either inside or outside the body. Natural virucides are produced by some plants such as eucalyptus and Australian tea trees.
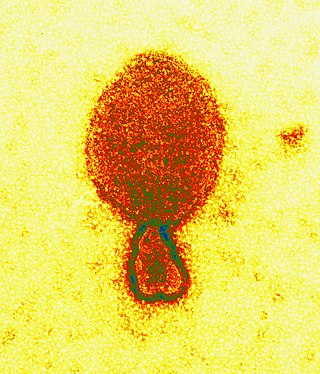
Henipavirus is a genus of negative-strand RNA viruses in the family Paramyxoviridae, order Mononegavirales containing six established species, and numerous others still under study. Henipaviruses are naturally harboured by several species of small mammals, notably pteropid fruit bats, microbats of several species, and shrews. Henipaviruses are characterised by long genomes and a wide host range. Their recent emergence as zoonotic pathogens capable of causing illness and death in domestic animals and humans is a cause of concern.

Influenza A virus (IAV) is a pathogen that causes the flu in birds and some mammals, including humans. It is an RNA virus whose subtypes have been isolated from wild birds. Occasionally, it is transmitted from wild to domestic birds, and this may cause severe disease, outbreaks, or human influenza pandemics.

Orthomyxoviridae is a family of negative-sense RNA viruses. It includes seven genera: Alphainfluenzavirus, Betainfluenzavirus, Gammainfluenzavirus, Deltainfluenzavirus, Isavirus, Thogotovirus, and Quaranjavirus. The first four genera contain viruses that cause influenza in birds and mammals, including humans. Isaviruses infect salmon; the thogotoviruses are arboviruses, infecting vertebrates and invertebrates. The Quaranjaviruses are also arboviruses, infecting vertebrates (birds) and invertebrates (arthropods).
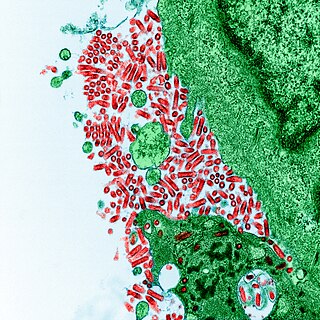
Rabies virus, scientific name Rabies lyssavirus, is a neurotropic virus that causes rabies in animals, including humans. Rabies transmission can occur through the saliva of animals and less commonly through contact with human saliva. Rabies lyssavirus, like many rhabdoviruses, has an extremely wide host range. In the wild it has been found infecting many mammalian species, while in the laboratory it has been found that birds can be infected, as well as cell cultures from mammals, birds, reptiles and insects. Rabies is reported in more than 150 countries and on all continents except Antarctica. The main burden of disease is reported in Asia and Africa, but some cases have been reported also in Europe in the past 10 years, especially in returning travellers.

Influenza C virus is the only species in the genus Gammainfluenzavirus, in the virus family Orthomyxoviridae, which like other influenza viruses, causes influenza.

Human Herpes Virus (HHV) Infected Cell Polypeptide 0 (ICP0) is a protein, encoded by the DNA of herpes viruses. It is produced by herpes viruses during the earliest stage of infection, when the virus has recently entered the host cell; this stage is known as the immediate-early or α ("alpha") phase of viral gene expression. During these early stages of infection, ICP0 protein is synthesized and transported to the nucleus of the infected host cell. Here, ICP0 promotes transcription from viral genes, disrupts structures in the nucleus known as nuclear dots or promyelocytic leukemia (PML) nuclear bodies, and alters the expression of host and viral genes in combination with a neuron specific protein. At later stages of cellular infection, ICP0 relocates to the cell cytoplasm to be incorporated into new virion particles.

H5N1 genetic structure is the molecular structure of the H5N1 virus's RNA.

Vincent R. Racaniello is a Higgins Professor in the Department of Microbiology and Immunology at Columbia University's College of Physicians and Surgeons. He is a co-author of a textbook on virology, Principles of Virology.
Yoshihiro Kawaoka is a virologist specializing in the study of the influenza and Ebola viruses. He holds a professorship in virology in the Department of Pathobiological Sciences at the University of Wisconsin-Madison, USA, and at the University of Tokyo, Japan.

Interferon regulatory factor 7, also known as IRF7, is a member of the interferon regulatory factor family of transcription factors.

Interferon-induced GTP-binding protein Mx1 is a protein that in humans is encoded by the MX1 gene.

Cleavage and polyadenylation specificity factor subunit 4 is a protein that in humans is encoded by the CPSF4 gene.

Influenza virus NS1A-binding protein is a protein that in humans is encoded by the IVNS1ABP gene.
Human bocavirus (HBoV) is the name given to all viruses in the genus Bocaparvovirus of virus family Parvoviridae that are known to infect humans. HBoV1 and HBoV3 are members of species Primate bocaparvovirus 1 whereas viruses HBoV2 and HBoV4 belong to species Primate bocaparvovirus 2. Some of these viruses cause human disease. HBoV1 is strongly implicated in causing some cases of lower respiratory tract infection, especially in young children, and several of the viruses have been linked to gastroenteritis, although the full clinical role of this emerging infectious disease remains to be elucidated.

Nonstructural protein 2 (NS2) is a viral protein found in the hepatitis C virus. It is also produced by influenza viruses, and is alternatively known as the nuclear export protein (NEP).
Adolfo García-Sastre,(born in Burgos, 10 October 1964) is a Spanish professor of Medicine and Microbiology and co-director of the Global Health & Emerging Pathogens Institute at the Icahn School of Medicine at Mount Sinai in New York City. His research into the biology of influenza viruses has been at the forefront of medical advances in epidemiology.

Peter Palese is a United States microbiologist, researcher, inventor and the Horace W. Goldsmith Professor in the Department of Microbiology at the Icahn School of Medicine at Mount Sinai in New York City, and an expert in the field of RNA viruses.
RIG-I-like receptors are a type of intracellular pattern recognition receptor involved in the recognition of viruses by the innate immune system. RIG-I is the best characterized receptor within the RIG-I like receptor (RLR) family. Together with MDA5 and LGP2, this family of cytoplasmic pattern recognition receptors (PRRs) are sentinels for intracellular viral RNA that is a product of viral infection. The RLR receptors provide frontline defence against viral infections in most tissues.
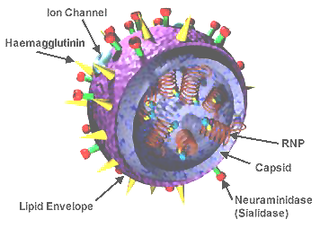
Influenza D virus is a species in the virus genus Deltainfluenzavirus, in the family Orthomyxoviridae, that causes influenza.
















