Related Research Articles
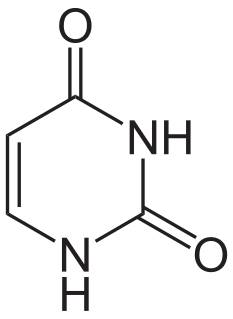
Uracil is one of the four nucleobases in the nucleic acid RNA that are represented by the letters A, G, C and U. The others are adenine (A), cytosine (C), and guanine (G). In RNA, uracil binds to adenine via two hydrogen bonds. In DNA, the uracil nucleobase is replaced by thymine. Uracil is a demethylated form of thymine.

Nucleobases, also known as nitrogenous bases or often simply bases, are nitrogen-containing biological compounds that form nucleosides, which, in turn, are components of nucleotides, with all of these monomers constituting the basic building blocks of nucleic acids. The ability of nucleobases to form base pairs and to stack one upon another leads directly to long-chain helical structures such as ribonucleic acid (RNA) and deoxyribonucleic acid (DNA).
The Nucleobase:Cation Symporter-1 (NCS1) Family (TC# 2.A.39) consists of over 1000 currently sequenced proteins derived from Gram-negative and Gram-positive bacteria, archaea, fungi and plants. These proteins function as transporters for nucleobases including purines and pyrimidines. Members of this family possess twelve transmembrane α-helical spanners (TMSs). At least some of them have been shown to function in uptake by substrate:H+ symport mechanism.
Proteins currently known to belong to the Ni2+-Co2+ Transporter (NiCoT) family (TC# 2.A.52) can be found in organisms ranging from Gram-negative and Gram-positive bacteria to archaea and some eukaryotes. Members of this family catalyze uptake of Ni2+ and/or Co2+ in a proton motive force-dependent process.
The amino acid-polyamine-organocation (APC) superfamily is the second largest superfamily of secondary carrier proteins currently known, and it contains several Solute carriers. Originally, the APC superfamily consisted of subfamilies under the transporter classification number. This superfamily has since been expanded to include eighteen different families.
The Amino Acid-Polyamine-Organocation (APC) Family of transport proteins includes members that function as solute:cation symporters and solute:solute antiporters. They occur in bacteria, archaea, fungi, unicellular eukaryotic protists, slime molds, plants and animals. They vary in length, being as small as 350 residues and as large as 850 residues. The smaller proteins are generally of prokaryotic origin while the larger ones are of eukaryotic origin. Most of them possess twelve transmembrane α-helical spanners but have a re-entrant loop involving TMSs 2 and 3. The APC Superfamily was established to encompass a wider range of homologues.
The bacterial murein precursor exporter (MPE) family is a member of the cation diffusion facilitator (CDF) superfamily of membrane transporters. Members of the MPE family are found in a large variety of Gram-negative and Gram-positive bacteria and facilitate the translocation of lipid-linked murein precursors. A representative list of proteins belonging to the MPE family can be found in the Transporter Classification Database.
The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) flippase superfamily is a group of integral membrane protein families. The MOP flippase superfamily includes twelve distantly related families, six for which functional data are available:
- One ubiquitous family (MATE) specific for drugs - (TC# 2.A.66.1) The Multi Antimicrobial Extrusion (MATE) Family
- One (PST) specific for polysaccharides and/or their lipid-linked precursors in prokaryotes - (TC# 2.A.66.2) The Polysaccharide Transport (PST) Family
- One (OLF) specific for lipid-linked oligosaccharide precursors of glycoproteins in eukaryotes - (TC# 2.A.66.3) The Oligosaccharidyl-lipid Flippase (OLF) Family
- One (MVF) lipid-peptidoglycan precursor flippase involved in cell wall biosynthesis - (TC# 2.A.66.4) The Mouse Virulence Factor (MVF) Family
- One (AgnG) which includes a single functionally characterized member that extrudes the antibiotic, Agrocin 84 - (TC# 2.A.66.5) The Agrocin 84 Antibiotic Exporter (AgnG) Family
- And finally, one (Ank) that shuttles inorganic pyrophosphate (PPi) - (TC# 2.A.66.9) The Progressive Ankylosis (Ank) Family
Lysine Exporters are a superfamily of transmembrane proteins which export amino acids, lipids and heavy metal ions. They provide ionic homeostasis, play a role in cell envelope assembly, and protect from excessive concentrations of heavy metals in cytoplasm. The superfamily was named based on the early discovery of the LysE carrier protein of Corynebacterium glutamicum.
The Disulfide bond oxidoreductase D (DsbD) family is a member of the Lysine Exporter (LysE) Superfamily. A representative list of proteins belonging to the DsbD family can be found in the Transporter Classification Base.
The gluconate:H+ symporter (GntP) family (TC# 2.A.8) is a family of transport proteins belonging to the ion transporter (IT) superfamily. Members of the GntP family include known gluconate permeases of E. coli and Bacillus species such as the D-Gluconate:H+ symporter of Bacillus subtillus (GntP; TC# 2.A.8.1.1) and the D-fructuronate/D-gluconate:H+ symporter of E. coli (GntP; TC# 2.A.8.1.3). A representative list of proteins belonging to the GntP family can be found in the Transporter Classification Database.
The Citrate-Mg2+:H+ (CitM) / Citrate-Ca2+:H+ (CitH) Symporter (CitMHS) Family (TC# 2.A.11) is a family of transport proteins belonging to the Ion transporter superfamily. Members of this family are found in Gram-positive and Gram-negative bacteria, archaea and possibly eukaryotes. These proteins all probably arose by an internal gene duplication event. Lensbouer & Doyle (2010) have reviewed these systems, classifying the porters with three superfamilies, according to ion-preference:
The arsenical resistance-3 (ACR3) family is a member of the BART superfamily. Based on operon analyses, ARC3 homologues may function either as secondary carriers or as primary active transporters, similarly to the ArsB and ArsAB families. In the latter case ATP hydrolysis again energizes transport. ARC3 homologues transport the same anions as ArsA/AB homologues, though ArsB homologues are members of the IT Superfamily and homologues of the ARC3 family are within the BART Superfamily suggesting they may not be evolutionarily related.
The NhaC family belongs to the Ion Transporter (IT) Superfamily. A representative list of proteins belonging to the NhaC family can be found in the Transporter Classification Database.
The Monovalent Cation:Proton Antiporter-2 (CPA2) Family is a moderately large family of transporters belonging to the CPA superfamily. Members of the CPA2 family have been found in bacteria, archaea and eukaryotes. The proteins of the CPA2 family consist of between 333 and 900 amino acyl residues and exhibit 10-14 transmembrane α-helical spanners (TMSs).
The Monovalent Cation (K+ or Na+):Proton Antiporter-3 (CPA3) Family (TC# 2.A.63) is a member of the Na+ transporting Mrp superfamily. The CPA3 family consists of bacterial multicomponent K+:H+ and Na+:H+ antiporters. The best characterized systems are the PhaABCDEFG system of Sinorhizobium meliloti (TC# 2.A.63.1.1) that functions in pH adaptation and as a K+ efflux system, and the MnhABCDEFG system of Staphylococcus aureus (TC# 2.A.63.1.3) that functions as a Na+ efflux Na+:H+ antiporter.
The Bacillus subtilis φ29 Holin Family is a group of transporters belonging to the Holin Superfamily IV. A representative list of members belonging to the φ29 holin family can be found in the Transporter Classification Database.
The Bacillus Spore Morphogenesis and Germination Holin (BSH) Family is a family of proteins named after a holin in Bacillus subtilis described to be involved in spore morphogenesis and germination by Real et al (2005). The gene encoding this holin is ywcE. Mutants lacking this gene or its product have spores that exhibit outer coat defects. These spores lack the characteristic striatal pattern resulting in the failure of the outer coat to attach to the underlying inner coat. Finally, the mutant spores accumulate reduced amounts of dipicolinic acid. BSH proteins average about 90 amino acyl residues in length and exhibit 3 transmembrane segments (TMSs). A representative list of homologous proteins, found only in Bacillus species, is available in the Transporter Classification Database.
The PTS Lactose-N,N’-Diacetylchitobiose (Lac) Family includes several sequenced lactose porters of Gram-positive bacteria, as well as the Escherichia coli and Borrelia burgdorferi N,N'-diacetylchitobiose (Chb) porters. It is part of the PTS-GFL superfamily. The former can transport aromatic β-glucosides and cellobiose, as well as Chb. However, only Chb induces expression of the chb operon.
The PTS L-Ascorbate (L-Asc) Family includes porters specific for L-ascorbate, and is part of the PTS-AG superfamily. A single PTS permease of the L-Asc family of PTS permeases has been functionally characterized. This is the SgaTBA system, renamed UlaABC by Yew and Gerlt.
References
- ↑ Karatza P, Panos P, Georgopoulou E, Frillingos S (Dec 2006). "Cysteine-scanning analysis of the nucleobase-ascorbate transporter signature motif in YgfO permease of Escherichia coli: Gln-324 and Asn-325 are essential, and Ile-329-Val-339 form an alpha-helix". The Journal of Biological Chemistry. 281 (52): 39881–90. doi: 10.1074/jbc.M605748200 . PMID 17077086.
- 1 2 Wong FH, Chen JS, Reddy V, Day JL, Shlykov MA, Wakabayashi ST, Saier MH (2012-01-01). "The amino acid-polyamine-organocation superfamily". Journal of Molecular Microbiology and Biotechnology. 22 (2): 105–13. doi: 10.1159/000338542 . PMID 22627175.
- ↑ Frillingos S (2012-01-01). "Insights to the evolution of Nucleobase-Ascorbate Transporters (NAT/NCS2 family) from the Cys-scanning analysis of xanthine permease XanQ". International Journal of Biochemistry and Molecular Biology. 3 (3): 250–72. PMC 3476789 . PMID 23097742.
- ↑ Diallinas G, Gournas C (2008-10-01). "Structure-function relationships in the nucleobase-ascorbate transporter (NAT) family: lessons from model microbial genetic systems". Channels. 2 (5): 363–72. doi: 10.4161/chan.2.5.6902 . PMID 18981714.
- ↑ Gournas C, Papageorgiou I, Diallinas G (May 2008). "The nucleobase-ascorbate transporter (NAT) family: genomics, evolution, structure-function relationships and physiological role". Molecular BioSystems. 4 (5): 404–16. doi:10.1039/b719777b. PMID 18414738.
- 1 2 3 Saier M Jr. "2.A.40 The Nucleobase/Ascorbate Transporter (NAT) or Nucleobase:Cation Symporter-2 (NCS2) Family". Transporter Classification Database. Saier Lab Bioinformatics Group / SDSC.
- 1 2 Lu F, Li S, Jiang Y, Jiang J, Fan H, Lu G, Deng D, Dang S, Zhang X, Wang J, Yan N (Apr 2011). "Structure and mechanism of the uracil transporter UraA". Nature. 472 (7342): 243–6. Bibcode:2011Natur.472..243L. doi:10.1038/nature09885. PMID 21423164. S2CID 4421922.
- ↑ Christiansen LC, Schou S, Nygaard P, Saxild HH (Apr 1997). "Xanthine metabolism in Bacillus subtilis: characterization of the xpt-pbuX operon and evidence for purine- and nitrogen-controlled expression of genes involved in xanthine salvage and catabolism". Journal of Bacteriology. 179 (8): 2540–50. doi:10.1128/jb.179.8.2540-2550.1997. PMC 179002 . PMID 9098051.
- ↑ Schultz AC, Nygaard P, Saxild HH (Jun 2001). "Functional analysis of 14 genes that constitute the purine catabolic pathway in Bacillus subtilis and evidence for a novel regulon controlled by the PucR transcription activator". Journal of Bacteriology. 183 (11): 3293–302. doi:10.1128/JB.183.11.3293-3302.2001. PMC 99626 . PMID 11344136.
- ↑ Ghim SY, Neuhard J (Jun 1994). "The pyrimidine biosynthesis operon of the thermophile Bacillus caldolyticus includes genes for uracil phosphoribosyltransferase and uracil permease". Journal of Bacteriology. 176 (12): 3698–707. doi:10.1128/jb.176.12.3698-3707.1994. PMC 205559 . PMID 8206848.
- ↑ Loh KD, Gyaneshwar P, Markenscoff Papadimitriou E, Fong R, Kim KS, Parales R, Zhou Z, Inwood W, Kustu S (Mar 2006). "A previously undescribed pathway for pyrimidine catabolism". Proceedings of the National Academy of Sciences of the United States of America. 103 (13): 5114–9. doi: 10.1073/pnas.0600521103 . PMC 1458803 . PMID 16540542.
- ↑ Kim KS, Pelton JG, Inwood WB, Andersen U, Kustu S, Wemmer DE (Aug 2010). "The Rut pathway for pyrimidine degradation: novel chemistry and toxicity problems". Journal of Bacteriology. 192 (16): 4089–102. doi:10.1128/JB.00201-10. PMC 2916427 . PMID 20400551.