
Brodmann area 9, or BA9, refers to a cytoarchitecturally defined portion of the frontal cortex in the brain of humans and other primates. Its cytoarchitecture is referred to as granular due to the concentration of granule cells in layer IV. It contributes to the dorsolateral and medial prefrontal cortex.
Neuroscience and intelligence refers to the various neurological factors that are partly responsible for the variation of intelligence within species or between different species. A large amount of research in this area has been focused on the neural basis of human intelligence. Historic approaches to studying the neuroscience of intelligence consisted of correlating external head parameters, for example head circumference, to intelligence. Post-mortem measures of brain weight and brain volume have also been used. More recent methodologies focus on examining correlates of intelligence within the living brain using techniques such as magnetic resonance imaging (MRI), functional MRI (fMRI), electroencephalography (EEG), positron emission tomography and other non-invasive measures of brain structure and activity.

The posterior cingulate cortex (PCC) is the caudal part of the cingulate cortex, located posterior to the anterior cingulate cortex. This is the upper part of the "limbic lobe". The cingulate cortex is made up of an area around the midline of the brain. Surrounding areas include the retrosplenial cortex and the precuneus.
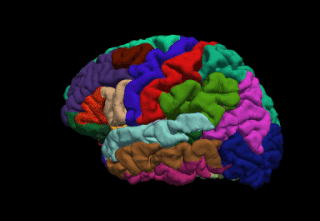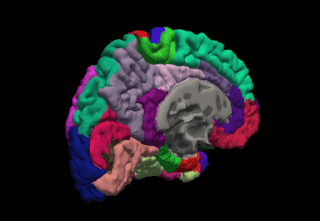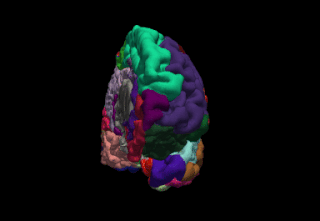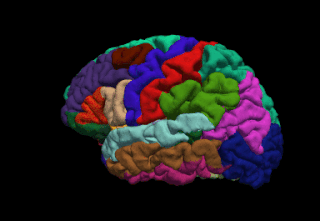
FreeSurfer is brain imaging software originally developed by Bruce Fischl, Anders Dale, Martin Sereno, and Doug Greve. Development and maintenance of FreeSurfer is now the primary responsibility of the Laboratory for Computational Neuroimaging at the Athinoula A. Martinos Center for Biomedical Imaging. FreeSurfer contains a set of programs with a common focus of analyzing magnetic resonance imaging (MRI) scans of brain tissue. It is an important tool in functional brain mapping and contains tools to conduct both volume based and surface based analysis. FreeSurfer includes tools for the reconstruction of topologically correct and geometrically accurate models of both the gray/white and pial surfaces, for measuring cortical thickness, surface area and folding, and for computing inter-subject registration based on the pattern of cortical folds.

Talairach coordinates, also known as Talairach space, is a 3-dimensional coordinate system of the human brain, which is used to map the location of brain structures independent from individual differences in the size and overall shape of the brain. It is still common to use Talairach coordinates in functional brain imaging studies and to target transcranial stimulation of brain regions. However, alternative methods such as the MNI Coordinate System have largely replaced Talairach for stereotaxy and other procedures.
The Organization for Human Brain Mapping (OHBM) is an organization of scientists with the main aim of organizing an annual meeting.

In neuroscience, the default mode network (DMN), also known as the default network, default state network, or anatomically the medial frontoparietal network (M-FPN), is a large-scale brain network primarily composed of the dorsal medial prefrontal cortex, posterior cingulate cortex, precuneus and angular gyrus. It is best known for being active when a person is not focused on the outside world and the brain is at wakeful rest, such as during daydreaming and mind-wandering. It can also be active during detailed thoughts related to external task performance. Other times that the DMN is active include when the individual is thinking about others, thinking about themselves, remembering the past, and planning for the future.
Alzheimer's Disease Neuroimaging Initiative (ADNI) is a multisite study that aims to improve clinical trials for the prevention and treatment of Alzheimer's disease (AD). This cooperative study combines expertise and funding from the private and public sector to study subjects with AD, as well as those who may develop AD and controls with no signs of cognitive impairment. Researchers at 63 sites in the US and Canada track the progression of AD in the human brain with neuroimaging, biochemical, and genetic biological markers. This knowledge helps to find better clinical trials for the prevention and treatment of AD. ADNI has made a global impact, firstly by developing a set of standardized protocols to allow the comparison of results from multiple centers, and secondly by its data-sharing policy which makes available all at the data without embargo to qualified researchers worldwide. To date, over 1000 scientific publications have used ADNI data. A number of other initiatives related to AD and other diseases have been designed and implemented using ADNI as a model. ADNI has been running since 2004 and is currently funded until 2021.
Anders Martin Dale is a prominent neuroscientist and professor of radiology, neurosciences, psychiatry, and cognitive science at the University of California, San Diego (UCSD), and is one of the world's leading developers of sophisticated computational neuroimaging techniques. He is the founding Director of the Center for Multimodal Imaging Genetics (CMIG) at UCSD.

Marcus E. Raichle is an American neurologist at the Washington University School of Medicine in Saint Louis, Missouri. He is a professor in the Department of Radiology with joint appointments in Neurology, Neurobiology and Biomedical Engineering. His research over the past 40 years has focused on the nature of functional brain imaging signals arising from PET and fMRI and the application of these techniques to the study of the human brain in health and disease. He received the Kavli Prize in Neuroscience “for the discovery of specialized brain networks for memory and cognition", together with Brenda Milner and John O’Keefe in 2014.

Resting state fMRI is a method of functional magnetic resonance imaging (fMRI) that is used in brain mapping to evaluate regional interactions that occur in a resting or task-negative state, when an explicit task is not being performed. A number of resting-state brain networks have been identified, one of which is the default mode network. These brain networks are observed through changes in blood flow in the brain which creates what is referred to as a blood-oxygen-level dependent (BOLD) signal that can be measured using fMRI.
Functional magnetic resonance spectroscopy of the brain (fMRS) uses magnetic resonance imaging (MRI) to study brain metabolism during brain activation. The data generated by fMRS usually shows spectra of resonances, instead of a brain image, as with MRI. The area under peaks in the spectrum represents relative concentrations of metabolites.

Russell "Russ" Alan Poldrack is an American psychologist and neuroscientist. He is a professor of psychology at Stanford University, associate director of Stanford Data Science, member of the Stanford Neuroscience Institute and director of the Stanford Center for Reproducible Neuroscience and the SDS Center for Open and Reproducible Science.

OpenNeuro is an open-science neuroinformatics database storing datasets from human brain imaging research studies.

The Brain Imaging Data Structure (BIDS) is a standard for organizing, annotating, and describing data collected during neuroimaging experiments. It is based on a formalized file and directory structure and metadata files with controlled vocabulary. This standard has been adopted by a multitude of labs around the world as well as databases such as OpenNeuro, SchizConnect, Developing Human Connectome Project, and FCP-INDI, and is seeing uptake in an increasing number of studies.

John-Dylan Haynes is a British-German brain researcher.
The occipital face area (OFA) is a region of the human cerebral cortex which is specialised for face perception. The OFA is located on the lateral surface of the occipital lobe adjacent to the inferior occipital gyrus. The OFA comprises a network of brain regions including the fusiform face area (FFA) and posterior superior temporal sulcus (STS) which support facial processing.

NeuroVault is an open-science neuroinformatics online repository of brain statistical maps atlases and parcellations.












