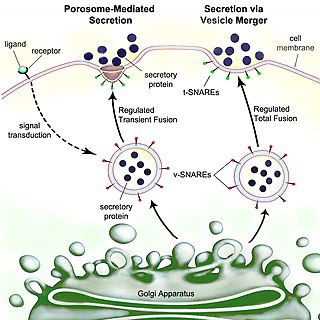
Secretion is the movement of material from one point to another, such as a secreted chemical substance from a cell or gland. In contrast, excretion is the removal of certain substances or waste products from a cell or organism. The classical mechanism of cell secretion is via secretory portals at the plasma membrane called porosomes. Porosomes are permanent cup-shaped lipoprotein structures embedded in the cell membrane, where secretory vesicles transiently dock and fuse to release intra-vesicular contents from the cell.
Braun's lipoprotein, found in some gram-negative cell walls, is one of the most abundant membrane proteins; its molecular weight is about 7.2 kDa. It is bound at its C-terminal end by a covalent bond to the peptidoglycan layer and is embedded in the outer membrane by its hydrophobic head. BLP tightly links the two layers and provides structural integrity to the outer membrane.

Yersinia pseudotuberculosis is a Gram-negative bacterium that causes Far East scarlet-like fever in humans, who occasionally get infected zoonotically, most often through the food-borne route. Animals are also infected by Y. pseudotuberculosis. The bacterium is urease positive.
Bacterial display is a protein engineering technique used for in vitro protein evolution. Libraries of polypeptides displayed on the surface of bacteria can be screened using flow cytometry or iterative selection procedures (biopanning). This protein engineering technique allows us to link the function of a protein with the gene that encodes it. Bacterial display can be used to find target proteins with desired properties and can be used to make affinity ligands which are cell-specific. This system can be used in many applications including the creation of novel vaccines, the identification of enzyme substrates and finding the affinity of a ligand for its target protein.
Virulence factors are cellular structures, molecules and regulatory systems that enable microbial pathogens to achieve the following:

A colicin is a type of bacteriocin produced by and toxic to some strains of Escherichia coli. Colicins are released into the environment to reduce competition from other bacterial strains. Colicins bind to outer membrane receptors, using them to translocate to the cytoplasm or cytoplasmic membrane, where they exert their cytotoxic effect, including depolarisation of the cytoplasmic membrane, DNase activity, RNase activity, or inhibition of murein synthesis.

The OmrA-B RNA gene family is a pair of homologous OmpR-regulated small non-coding RNA that was discovered in E. coli during two large-scale screens. OmrA-B is highly abundant in stationary phase, but low levels could be detected in exponentially growing cells as well. RygB is adjacent to RygA a closely related RNA. These RNAs bind to the Hfq protein and regulate gene expression by antisense binding. They negatively regulate the expression of several genes encoding outer membrane proteins, including cirA, CsgD, fecA, fepA and ompT by binding in the vicinity of the Shine-Dalgarno sequence, suggesting the control of these targets is dependent on Hfq protein and RNase E. Taken together, these data suggest that OmrA-B participates in the regulation of outer membrane composition, responding to environmental conditions.

The micF RNA is a non-coding RNA stress response gene found in Escherichia coli and related bacteria that post-transcriptionally controls expression of the outer membrane porin gene ompF. The micF gene encodes a non-translated 93 nucleotide antisense RNA that binds its target ompF mRNA and regulates ompF expression by inhibiting translation and inducing degradation of the message. In addition, other factors, such as the RNA chaperone protein StpA also play a role in this regulatory system. The expression of micF is controlled by both environmental and internal stress factors. Four transcriptional regulators are known to bind the micF promoter region and activate micF expression.

General bacterial porins are a family of porin proteins from the outer membranes of Gram-negative bacteria. The porins act as molecular filters for hydrophilic compounds. They are responsible for the 'molecular sieve' properties of the outer membrane. Porins form large water-filled channels which allow the diffusion of hydrophilic molecules into the periplasmic space. Some porins form general diffusion channels that allow any solute up to a certain size to cross the membrane, while other porins are specific for one particular solute and contain a binding site for that solute inside the pores. As porins are the major outer membrane proteins, they also serve as receptor sites for the binding of phages and bacteriocins.

Virulence-related outer membrane proteins, or outer surface proteins (Osp) in some contexts, are expressed in the outer membrane of gram-negative bacteria and are essential to bacterial survival within macrophages and for eukaryotic cell invasion.

VrrA is a non-coding RNA that is conserved across all Vibrio species of bacteria and acts as a repressor for the synthesis of the outer membrane protein OmpA. This non-coding RNA was initially identified from Tn5 transposon mutant libraries of Vibrio cholerae and its location within the bacterial genome was mapped to the intergenic region between genes VC1741 and VC1743 by RACE analysis.
The RTX toxin superfamily is a group of cytolysins and cytotoxins produced by bacteria. There are over 1000 known members with a variety of functions. The RTX family is defined by two common features: characteristic repeats in the toxin protein sequences, and extracellular secretion by the type I secretion systems (T1SS). The name RTX refers to the glycine and aspartate-rich repeats located at the C-terminus of the toxin proteins, which facilitate export by a dedicated T1SS encoded within the rtx operon.
Bacterial small RNAs (bsRNA) are small RNAs produced by bacteria; they are 50- to 500-nucleotide non-coding RNA molecules, highly structured and containing several stem-loops. Numerous sRNAs have been identified using both computational analysis and laboratory-based techniques such as Northern blotting, microarrays and RNA-Seq in a number of bacterial species including Escherichia coli, the model pathogen Salmonella, the nitrogen-fixing alphaproteobacterium Sinorhizobium meliloti, marine cyanobacteria, Francisella tularensis, Streptococcus pyogenes, the pathogen Staphylococcus aureus, and the plant pathogen Xanthomonas oryzae pathovar oryzae. Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNA or protein, and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress response, and structure.
Omptins are a family of bacterial proteases. They are aspartate proteases, which cleave peptides with the use of a water molecule. Found in the outer membrane of gram-negative enterobacteria such as Shigella flexneri, Yersinia pestis, Escherichia coli, and Salmonella enterica. Omptins consist of a widely conserved beta barrel spanning the membrane with 5 extracellular loops. These loops are responsible for the various substrate specificities. These proteases rely upon binding of lipopolysaccharide for activity.
EnvZ/OmpR is a two-component regulatory system widely distributed in bacteria and particularly well characterized in Escherichia coli. Its function is in osmoregulation, responding to changes in environmental osmolality by regulating the expression of the outer membrane porins OmpF and OmpC. EnvZ is a histidine kinase which also possesses a cytoplasmic osmosensory domain, and OmpR is its corresponding response regulator protein.

In molecular biology, the OmpA domain is a conserved protein domain with a beta/alpha/beta/alpha-beta(2) structure found in the C-terminal region of many Gram-negative bacterial outer membrane proteins, such as porin-like integral membrane proteins, small lipid-anchored proteins, and MotB proton channels. The N-terminal half of these proteins is variable although some of the proteins in this group have the OmpA-like transmembrane domain at the N terminus. OmpA from Escherichia coli is required for pathogenesis, and can interact with host receptor molecules. MotB serve two functions in E. coli, the MotA(4)-MotB(2) complex attaches to the cell wall via MotB to form the stator of the flagellar motor, and the MotA-MotB complex couples the flow of ions across the cell membrane to movement of the rotor.

In molecular biology, trimeric autotransporter adhesins (TAAs), are proteins found on the outer membrane of Gram-negative bacteria. Bacteria use TAAs in order to infect their host cells via a process called cell adhesion. TAAs also go by another name, oligomeric coiled-coil adhesins, which is shortened to OCAs. In essence, they are virulence factors, factors that make the bacteria harmful and infective to the host organism.

In molecular biology, YadA is a protein domain which is short for Yersinia adhesin A. These proteins have strong sequence and structural homology, particularly at their C-terminal end. The function is to promote their pathogenicity and virulence in host cells, though cell adhesion. YadA is found in three pathogenic species of Yersinia, Y. pestis,Y. pseudotuberculosis, and Y. enterocolitica. The YadA domain is encoded for by a virulence plasmid in Yersinia, which encodes a type-III secretion (T3S) system consisting of the Ysc injectisome and the Yop effectors.

OmpT is an aspartyl protease found on the outer membrane of Escherichia coli. OmpT is a subtype of the family of omptin proteases, which are found on some gram-negative species of bacteria.

Bacterial secretion systems are protein complexes present on the cell membranes of bacteria for secretion of substances. Specifically, they are the cellular devices used by pathogenic bacteria to secrete their virulence factors to invade the host cells. They can be classified into different types based on their specific structure, composition and activity. Generally, proteins can be secreted through two different processes. One process is a one-step mechanism in which proteins from the cytoplasm of bacteria are transported and delivered directly through the cell membrane into the host cell. Another involves a two-step activity in which the proteins are first transported out of the inner cell membrane, then deposited in the periplasm, and finally through the outer cell membrane into the host cell.













