
Schizosaccharomyces pombe, also called "fission yeast", is a species of yeast used in traditional brewing and as a model organism in molecular and cell biology. It is a unicellular eukaryote, whose cells are rod-shaped. Cells typically measure 3 to 4 micrometres in diameter and 7 to 14 micrometres in length. Its genome, which is approximately 14.1 million base pairs, is estimated to contain 4,970 protein-coding genes and at least 450 non-coding RNAs.

Biological databases are libraries of biological sciences, collected from scientific experiments, published literature, high-throughput experiment technology, and computational analysis. They contain information from research areas including genomics, proteomics, metabolomics, microarray gene expression, and phylogenetics. Information contained in biological databases includes gene function, structure, localization, clinical effects of mutations as well as similarities of biological sequences and structures.
In genetics, an expressed sequence tag (EST) is a short sub-sequence of a cDNA sequence. ESTs may be used to identify gene transcripts, and are instrumental in gene discovery and in gene-sequence determination. The identification of ESTs has proceeded rapidly, with approximately 74.2 million ESTs now available in public databases.

The origin of replication is a particular sequence in a genome at which replication is initiated. Propagation of the genetic material between generations requires timely and accurate duplication of DNA by semiconservative replication prior to cell division to ensure each daughter cell receives the full complement of chromosomes. This can either involve the replication of DNA in living organisms such as prokaryotes and eukaryotes, or that of DNA or RNA in viruses, such as double-stranded RNA viruses. Synthesis of daughter strands starts at discrete sites, termed replication origins, and proceeds in a bidirectional manner until all genomic DNA is replicated. Despite the fundamental nature of these events, organisms have evolved surprisingly divergent strategies that control replication onset. Although the specific replication origin organization structure and recognition varies from species to species, some common characteristics are shared.
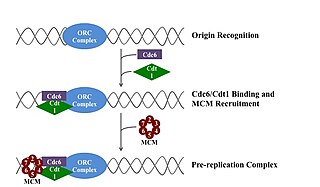
A pre-replication complex (pre-RC) is a protein complex that forms at the origin of replication during the initiation step of DNA replication. Formation of the pre-RC is required for DNA replication to occur. Complete and faithful replication of the genome ensures that each daughter cell will carry the same genetic information as the parent cell. Accordingly, formation of the pre-RC is a very important part of the cell cycle.

Ensembl genome database project is a scientific project at the European Bioinformatics Institute, which was launched in 1999 in response to the imminent completion of the Human Genome Project. Ensembl aims to provide a centralized resource for geneticists, molecular biologists and other researchers studying the genomes of our own species and other vertebrates and model organisms. Ensembl is one of several well known genome browsers for the retrieval of genomic information.
The DrugBank database is a comprehensive, freely accessible, online database containing information on drugs and drug targets. As both a bioinformatics and a cheminformatics resource, DrugBank combines detailed drug data with comprehensive drug target information. DrugBank has used content from Wikipedia; Wikipedia also often links to Drugbank, posing potential circular reporting issues.

The Biological General Repository for Interaction Datasets (BioGRID) is a curated biological database of protein-protein interactions, genetic interactions, chemical interactions, and post-translational modifications created in 2003 (originally referred to as simply the General Repository for Interaction Datasets by Mike Tyers, Bobby-Joe Breitkreutz, and Chris Stark at the Lunenfeld-Tanenbaum Research Institute at Mount Sinai Hospital. It strives to provide a comprehensive curated resource for all major model organism species while attempting to remove redundancy to create a single mapping of data. Users of The BioGRID can search for their protein, chemical or publication of interest and retrieve annotation, as well as curated data as reported, by the primary literature and compiled by in house large-scale curation efforts. The BioGRID is hosted in Toronto, Ontario, Canada and Dallas, Texas, United States and is partnered with the Saccharomyces Genome Database, FlyBase, WormBase, PomBase, and the Alliance of Genome Resources. The BioGRID is funded by the NIH and CIHR. BioGRID is an observer member of the International Molecular Exchange Consortium.
The Saccharomyces Genome Database (SGD) is a scientific database of the molecular biology and genetics of the yeast Saccharomyces cerevisiae, which is commonly known as baker's or budding yeast.

DNA polymerase epsilon subunit 2 is an enzyme that in humans is encoded by the POLE2 gene.
BIOBASE is an international bioinformatics company headquartered in Wolfenbüttel, Germany. The company focuses on the generation, maintenance, and licensing of databases in the field of molecular biology, and their related software platforms.
ChimerDB is a database of fusion sequences.
RegulonDB is a database of the regulatory network of gene expression in Escherichia coli K-12. RegulonDB also models the organization of the genes in transcription units, operons and regulons. A total of 120 sRNAs with 231 total interactions which all together regulate 192 genes are also included. RegulonDB was founded in 1998 and also contributes data to the EcoCyc database.
YEASTRACT is a curated repository of more than 48000 regulatory associations between transcription factors (TF) and target genes in Saccharomyces cerevisiae, based on more than 1200 bibliographic references. It also includes the description of about 300 specific DNA binding sites for more than a hundred characterized TFs. Further information about each Yeast gene has been extracted from the Saccharomyces Genome Database (SGD). For each gene the associated Gene Ontology (GO) terms and their hierarchy in GO was obtained from the GO consortium. Currently, YEASTRACT maintains more than 7100 terms from GO. The nucleotide sequences of the promoter and coding regions for Yeast genes were obtained from Regulatory Sequence Analysis Tools (RSAT). All the information in YEASTRACT is updated regularly to match the latest data from SGD, GO consortium, RSA Tools and recent literature on yeast regulatory networks.
LocDB is an expert-curated database that collects experimental annotations for the subcellular localization of proteins in Homo sapiens (human) and Arabidopsis thaliana (Weed). The database also contains predictions of subcellular localization from a variety of state-of-the-art prediction methods for all proteins with experimental information.

PSORTdb is a database of protein subcellular localization (SCL) for bacteria and archaea. It is a member of the PSORT family of bioinformatics tools. The database consists of two datasets, ePSORTdb and cPSORTdb, which contain information determined through experimental validation and computational prediction, respectively. The ePSORTdb dataset is the largest curated collection of experimentally verified SCL data.

The Human Metabolome Database (HMDB) is a comprehensive, high-quality, freely accessible, online database of small molecule metabolites found in the human body. Created by the Human Metabolome Project funded by Genome Canada. One of the first dedicated metabolomics databases, the HMDB facilitates human metabolomics research, including the identification and characterization of human metabolites using NMR spectroscopy, GC-MS spectrometry and LC/MS spectrometry. To aid in this discovery process, the HMDB contains three kinds of data: 1) chemical data, 2) clinical data, and 3) molecular biology/biochemistry data. The chemical data includes 41,514 metabolite structures with detailed descriptions along with nearly 10,000 NMR, GC-MS and LC/MS spectra.
MetaboAnalyst is a set of online tools for metabolomic data analysis and interpretation, created by members of the Wishart Research Group at the University of Alberta. It was first released in May 2009 and version 2.0 was released in January 2012. MetaboAnalyst provides a variety of analysis methods that have been tailored for metabolomic data. These methods include metabolomic data processing, normalization, multivariate statistical analysis, and data annotation. The current version is focused on biomarker discovery and classification.
PomBase is a model organism database that provides online access to the fission yeast Schizosaccharomyces pombe genome sequence and annotated features, together with a wide range of manually curated functional gene-specific data. The PomBase website was redeveloped in 2016 to provide users with a more fully integrated, better-performing service.








