
β-Galactosidase, is a glycoside hydrolase enzyme that catalyzes the following process:

Aspartate carbamoyltransferase catalyzes the first step in the pyrimidine biosynthetic pathway.
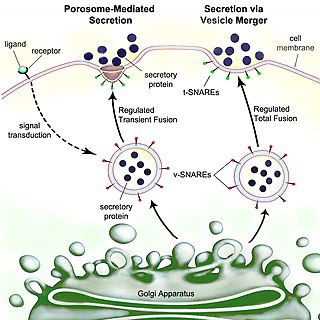
Secretion is the movement of material from one point to another, such as a secreted chemical substance from a cell or gland. In contrast, excretion is the removal of certain substances or waste products from a cell or organism. The classical mechanism of cell secretion is via secretory portals at the plasma membrane called porosomes. Porosomes are permanent cup-shaped lipoprotein structures embedded in the cell membrane, where secretory vesicles transiently dock and fuse to release intra-vesicular contents from the cell.

Porins are beta barrel proteins that cross a cellular membrane and act as a pore, through which molecules can diffuse. Unlike other membrane transport proteins, porins are large enough to allow passive diffusion, i.e., they act as channels that are specific to different types of molecules. They are present in the outer membrane of gram-negative bacteria and some gram-positive mycobacteria, the outer membrane of mitochondria, and the outer chloroplast membrane.
PEP group translocation, also known as the phosphotransferase system or PTS, is a distinct method used by bacteria for sugar uptake where the source of energy is from phosphoenolpyruvate (PEP). It is known to be a multicomponent system that always involves enzymes of the plasma membrane and those in the cytoplasm.
Translocase is a general term for a protein that assists in moving another molecule, usually across a cell membrane. These enzymes catalyze the movement of ions or molecules across membranes or their separation within membranes. The reaction is designated as a transfer from “side 1” to “side 2” because the designations “in” and “out”, which had previously been used, can be ambiguous. Translocases are the most common secretion system in Gram positive bacteria.

Cystathionine beta-lyase, also commonly referred to as CBL or β-cystathionase, is an enzyme that primarily catalyzes the following α,β-elimination reaction

General bacterial porins are a family of porin proteins from the outer membranes of Gram-negative bacteria. The porins act as molecular filters for hydrophilic compounds. They are responsible for the 'molecular sieve' properties of the outer membrane. Porins form large water-filled channels which allow the diffusion of hydrophilic molecules into the periplasmic space. Some porins form general diffusion channels that allow any solute up to a certain size to cross the membrane, while other porins are specific for one particular solute and contain a binding site for that solute inside the pores. As porins are the major outer membrane proteins, they also serve as receptor sites for the binding of phages and bacteriocins.

Outer membrane receptors, also known as TonB-dependent receptors, are a family of beta barrel proteins named for their localization in the outer membrane of gram-negative bacteria. TonB complexes sense signals from the outside of bacterial cells and transmit them into the cytoplasm, leading to transcriptional activation of target genes. TonB-dependent receptors in gram-negative bacteria are associated with the uptake and transport of large substrates such as iron siderophore complexes and vitamin B12.
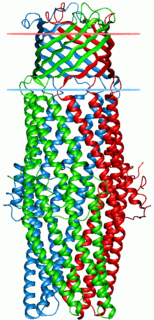
Proteins in the outer membrane efflux protein family form trimeric (three-piece) channels that allow export of a variety of substrates in gram-negative bacteria. Each member of this family is composed of two repeats. The trimeric channel is composed of a 12-stranded beta-barrel that spans the outer membrane, and a long all helical barrel that spans the periplasm.
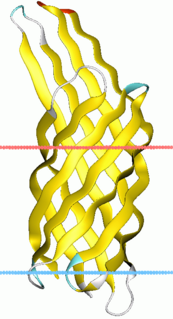
Virulence-related outer membrane proteins, or outer surface proteins (Osp) in some contexts, are expressed in the outer membrane of gram-negative bacteria and are essential to bacterial survival within macrophages and for eukaryotic cell invasion.

The fimbrial usher protein is involved in biogenesis of the pilus in Gram-negative bacteria. The biogenesis of some fimbriae requires a two-component assembly and transport system which is composed of a periplasmic chaperone and a pore-forming outer membrane protein which has been termed a molecular 'usher'; this is the chaperone-usher pathway.
Many bacteria secrete small iron-binding molecules called siderophores, which bind strongly to ferric ions. FepA is an integral bacterial outer membrane porin protein that belongs to outer membrane receptor family and provides the active transport of iron bound by the siderophore enterobactin from the extracellular space, into the periplasm of Gram-negative bacteria. FepA has also been shown to transport vitamin B12, and colicins B and D as well. This protein belongs to family of ligand-gated protein channels.

The Escherichia coliAcriflavine resistance encode a multi-drug efflux system that is believed to protect the bacterium against hydrophobic inhibitors. The E. coli AcrB protein is a transporter that is energized by proton-motive force and that shows the widest substrate specificity among all known multidrug pumps, ranging from most of the currently used antibiotics, disinfectants, dyes, and detergents to simple solvents.

In molecular biology, YadA is a protein domain which is short for Yersinia adhesin A. These proteins have strong sequence and structural homology, particularly at their C-terminal end. The function is to promote their pathogenicity and virulence in host cells, though cell adhesion. YadA is found in three pathogenic species of Yersinia, Y. pestis,Y. pseudotuberculosis, and Y. enterocolitica. The YadA domain is encoded for by a virulence plasmid in Yersinia, which encodes a type-III secretion (T3S) system consisting of the Ysc injectisome and the Yop effectors.

OmpT is an aspartyl protease found on the outer membrane of Escherichia coli. OmpT is a subtype of the family of omptin proteases, which are found on some gram-negative species of bacteria.
Chaperone-usher fimbriae (CU) are linear, unbranching, outer-membrane pili secreted by gram-negative bacteria through the chaperone-usher system rather than through type IV secretion or extracellular nucleation systems. These fimbriae are built up out of modular pilus subunits, which are transported into the periplasm in a Sec dependent manner. Chaperone-usher secreted fimbriae are important pathogenicity factors facilitating host colonisation, localisation and biofilm formation in clinically important species such as uropathogenic Escherichia coli and Pseudomonas aeruginosa.
Gabriel Waksman FMedSci, FRS, is Courtauld professor of biochemistry and molecular biology at University College London (UCL), and professor of structural and molecular biology at Birkbeck College, University of London. He is the director of the Institute of Structural and Molecular Biology (ISMB) at UCL and Birkbeck, head of the Department of Structural and Molecular Biology at UCL, and head of the Department of Biological Sciences at Birkbeck.

Resistance-nodulation-division (RND) family transporters are a category of bacterial efflux pumps, especially identified in Gram-negative bacteria and located in the cytoplasmic membrane, that actively transport substrates. The RND superfamily includes seven families: the heavy metal efflux (HME), the hydrophobe/amphiphile efflux-1, the nodulation factor exporter family (NFE), the SecDF protein-secretion accessory protein family, the hydrophobe/amphiphile efflux-2 family, the eukaryotic sterol homeostasis family, and the hydrophobe/amphiphile efflux-3 family. These RND systems are involved in maintaining homeostasis of the cell, removal of toxic compounds, and export of virulence determinants. They have a broad substrate spectrum and can lead to the diminished activity of unrelated drug classes if over-expressed. The first reports of drug resistant bacterial infections were reported in the 1940s after the first mass production of antibiotics. Most of the RND superfamily transport systems are made of large polypeptide chains. RND proteins exist primarily in gram-negative bacteria but can also be found in gram-positive bacteria, archaea, and eukaryotes.
Contact-dependent growth inhibition (CDI) is a phenomenon where a bacterial cell may deliver a polymorphic toxin molecule into neighbouring bacterial cells upon direct cell-cell contact, causing growth arrest or cell death.














