Function
PRC1 protein is expressed at relatively high levels during S and G2/M phases of the cell cycle before dropping dramatically after mitotic exit and entrance into G1 phase. PRC1 is located in the nucleus during interphase, becomes associated with the mitotic spindle in a highly dynamic manner during anaphase, and localizes to the cell midbody during cytokinesis. PRC1 was first identified in 1998 using an ''in vitro'' phosphorylation screening method and shown to be a substrate of several cyclin-dependent kinases (CDKs). [5] Correspondingly, ablation of PRC1 has been shown to disrupt spindle midzone assembly in mammalian systems. [7]
At least three alternatively spliced transcript variants encoding distinct isoforms of PRC1 have been observed. [6] Additionally, PRC1 has sequence homology with Ase1 in yeasts, SPD-1 (spindle defective 1) in C. elegans, Feo in D. melanogaster, and MAP65 in plants, all of which fall in a conserved family of nonmotor microtubule-associated proteins (MAPs). [8] [9] [10]
Structure
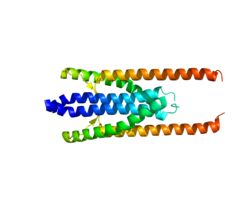
The crystal structure of PRC1 has only recently been characterized in vitro. In 2013, PRC1 was illustrated as a lengthy molecule consisting of a C-terminal spectrin microtubule-binding domain, an extended rod domain, and an N-terminal dimerization domain. [9] [11] Consisting of an intricate arrangement of α-helices, the rod domain, together with the dimerization-conducting N terminus cooperate to facilitate binding of other proteins, such as Kinesin-4, to PRC1. PRC1’s rod domain adopts multiple conformations, all affected by its C-terminal spectrin domain. A model has been suggested in which PRC1 is likely to be a flexible molecule both in solution and on single microtubules but becomes more rigid when the microtubule-binding domains are restricted with antiparallel microtubule filament crosslinking, seen at the spindle midzone. The overall structure of the PRC1 homodimer is reminiscent of actin-bundling proteins, and this process of microtubule filament crosslinking is similar to that of actin. [9]
Role in cytokinesis
PRC1’s role in midzone microtubule formation, essential to the cytokinetic machinery of mammals, is made possible through its collaboration with Kinesin-4 in setting up a controlled zone of overlapping, antiparallel microtubules at the spindle midzone. [12] PRC1 is normally inhibited until anaphase onset by CDK1 mediated phosphorylation, preventing its dimerization. Upon anaphase onset and removal of inhibitory CDK1 phosphorylation, PRC1 dimers form. These homodimers specifically recognize antiparallel microtubule overlaps, found at the spindle midzone, and bind, allowing microtubule sliding, cross-linking of microtubule filaments, and assembly of central-spindle-mediating proteins, including but not limited to Kinesin-4. [12] [13]
PRC1 dimers, required for the high-affinity interaction with Kinesin-4, recruit Kinesin-4 to regions of antiparallel microtubule overlap, where Kinesin-4, a plus-end directed motor protein that inhibits microtubule dynamics, helps to form length-dependent end tags that help stabilize and regulate spindle microtubule assembly within cytokinesis. [9] [12] This PRC1-Kinesin-4 complex differentially identifies and regulates the spindle midzone microtubules during cell division. [12] This regulation is crucial in order for cytokinesis to progress properly.
This page is based on this
Wikipedia article Text is available under the
CC BY-SA 4.0 license; additional terms may apply.
Images, videos and audio are available under their respective licenses.