Building
In general there are two different data types that are used in the construction of a phylogenetic tree. In distance-based computations a phylogenetic tree is created by analyzing relationships among the distance between species and the edge lengths of a corresponding tree. Using a character-based approach employs character states across species as an input in an attempt to find the most "perfect" phylogenetic tree. [3] [4]
The statistical components of a perfect phylogenetic tree can best be described as follows: [3]
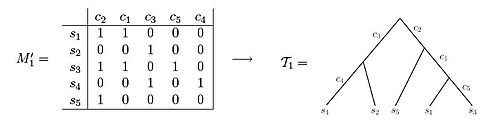
A perfect phylogeny for an n x m character state matrix M is a rooted tree T with n leaves satisfying:
i. Each row of M labels exactly one leaf of T
ii. Each column of M labels exactly one edge of T
iii. Every interior edge of T is labeled by at least one column of M
iv. The characters associated with the edges along the unique path from root to a leaf v exactly specify the character vector of v, i.e. the character vector has a 1 entry in all columns corresponding to characters associated to path edges and a 0 entry otherwise.
It is worth noting that it is very rare to find actual phylogenetic data that adheres to the concepts and limitations detailed here. Therefore, it is often the case that researchers are forced to compromise by developing trees that simply try to minimize homoplasy, finding a maximum-cardinality set of compatible characters, or constructing phylogenies that match as closely as possible to the partitions implied by the characters.
Usage
Perfect phylogeny is a theoretical framework that can also be used in more practical methods. One such example is that of Incomplete Directed Perfect Phylogeny. This concept involves utilizing perfect phylogenies with real, and therefore incomplete and imperfect, datasets. Such a method utilizes SINEs to determine evolutionary similarity. These Short Interspersed Elements are present across many genomes and can be identified by their flanking sequences. SINEs provide information on the inheritance of certain traits across different species. Unfortunately, if a SINE is missing it is difficult to know whether those SINEs were present prior to the deletion. By utilizing algorithms derived from perfect phylogeny data we are able to attempt to reconstruct a phylogenetic tree in spite of these limitations. [10]
Perfect phylogeny is also used in the construction of haplotype maps. By utilizing the concepts and algorithms described in perfect phylogeny one can determine information regarding missing and unavailable haplotype data. [11] By assuming that the set of haplotypes that result from genotype mapping corresponds and adheres to the concept of perfect phylogeny (as well as other assumptions such as perfect Mendelian inheritance and the fact that there is only one mutation per SNP), one is able to infer missing haplotype data. [12] [13] [14] [15]
Inferring a phylogeny from noisy VAF data under the PPM is a hard problem. [5] Most inference tools include some heuristic step to make inference computationally tractable. Examples of tools that infer phylogenies from noisy VAF data include AncesTree, Canopy, CITUP, EXACT, and PhyloWGS. [5] [6] [7] [8] [9] In particular, EXACT performs exact inference by using GPUs to compute a posterior probability on all possible trees for small size problems. Extensions to the PPM have been made with accompanying tools. [16] [17] For example, tools such as MEDICC, TuMult, and FISHtrees allow the number of copies of a given genetic element, or ploidy, to both increase, or decrease, thus effectively allowing the removal of mutations. [18] [19] [20]
This page is based on this
Wikipedia article Text is available under the
CC BY-SA 4.0 license; additional terms may apply.
Images, videos and audio are available under their respective licenses.