
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers – specifically polypeptides – formed from sequences of amino acids, the monomers of the polymer. A single amino acid monomer may also be called a residue indicating a repeating unit of a polymer. Proteins form by amino acids undergoing condensation reactions, in which the amino acids lose one water molecule per reaction in order to attach to one another with a peptide bond. By convention, a chain under 30 amino acids is often identified as a peptide, rather than a protein. To be able to perform their biological function, proteins fold into one or more specific spatial conformations driven by a number of non-covalent interactions such as hydrogen bonding, ionic interactions, Van der Waals forces, and hydrophobic packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their three-dimensional structure. This is the topic of the scientific field of structural biology, which employs techniques such as X-ray crystallography, NMR spectroscopy, cryo electron microscopy (cryo-EM) and dual polarisation interferometry to determine the structure of proteins.

In molecular biology, an intrinsically disordered protein (IDP) is a protein that lacks a fixed or ordered three-dimensional structure, typically in the absence of its macromolecular interaction partners, such as other proteins or RNA. IDPs range from fully unstructured to partially structured and include random coil, molten globule-like aggregates, or flexible linkers in large multi-domain proteins. They are sometimes considered as a separate class of proteins along with globular, fibrous and membrane proteins.

An ATP-binding motif is a 250-residue sequence within an ATP-binding protein’s primary structure. The binding motif is associated with a protein’s structure and/or function. ATP is a molecule of energy, and can be a coenzyme, involved in a number of biological reactions. ATP is proficient at interacting with other molecules through a binding site. The ATP binding site is the environment in which ATP catalytically actives the enzyme and, as a result, is hydrolyzed to ADP. The binding of ATP causes a conformational change to the enzyme it is interacting with.

John Kuriyan is the Dean of Basic Sciences and a Professor of Biochemistry at Vanderbilt University School of Medicine. He was formerly the Chancellor's Professor at the University of California, Berkeley in the departments of Molecular and Cell Biology (MCB) and Chemistry, a Faculty Scientist in Berkeley Lab's Physical Biosciences Division, and a Howard Hughes Medical Institute investigator. He is a member of the National Academy of Sciences and he has also been on the Life Sciences jury for the Infosys Prize in 2009, 2019 and 2020.

Proteins are generally thought to adopt unique structures determined by their amino acid sequences. However, proteins are not strictly static objects, but rather populate ensembles of conformations. Transitions between these states occur on a variety of length scales and time scales, and have been linked to functionally relevant phenomena such as allosteric signaling and enzyme catalysis.
Dorothee Kern, is a professor of Biochemistry at Brandeis University and former player for the East German national basketball team.

Paul Reinhard Schimmel is an American biophysical chemist and translational medicine pioneer.

Cubilin is a protein that in humans is encoded by the CUBN gene.
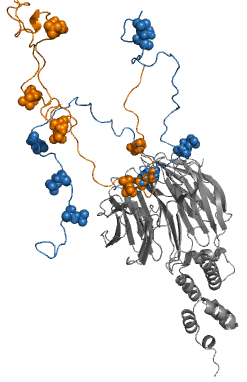
Fuzzy complexes are protein complexes, where structural ambiguity or multiplicity exists and is required for biological function. Alteration, truncation or removal of conformationally ambiguous regions impacts the activity of the corresponding complex. Fuzzy complexes are generally formed by intrinsically disordered proteins. Structural multiplicity usually underlies functional multiplicity of protein complexes following a fuzzy logic. Distinct binding modes of the nucleosome are also regarded as a special case of fuzziness.

Yoshinori Ohsumi is a Japanese cell biologist specializing in autophagy, the process that cells use to destroy and recycle cellular components. Ohsumi is a professor at Tokyo Institute of Technology's Institute of Innovative Research. He received the Kyoto Prize for Basic Sciences in 2012, the 2016 Nobel Prize in Physiology or Medicine, and the 2017 Breakthrough Prize in Life Sciences for his discoveries of mechanisms for autophagy.

In computational chemistry, conformational ensembles, also known as structural ensembles, are experimentally constrained computational models describing the structure of intrinsically unstructured proteins. Such proteins are flexible in nature, lacking a stable tertiary structure, and therefore cannot be described with a single structural representation. The techniques of ensemble calculation are relatively new on the field of structural biology, and are still facing certain limitations that need to be addressed before it will become comparable to classical structural description methods such as biological macromolecular crystallography.
Repeat Associated Non-AUG translation, or RAN translation, is an irregular mode of mRNA translation that can occur in eukaryotic cells.
Eric M. Verdin is the 5th President and Chief Executive Officer of the Buck Institute for Research on Aging. He is a native of Belgium. Dr. Verdin is also a Professor of Medicine at University of California, San Francisco.

Helen Jane Dyson is a British-born biophysicist and a professor of integrative structural and computational biology at the Scripps Research Institute in La Jolla, California. She also serves as editor-in-chief of the Biophysical Journal. She was elected a Member of the National Academy of Sciences in 2022.
In molecular biology, an arginine finger is an amino acid residue of some enzymes. Arginine fingers are often found in the protein superfamily of AAA+ ATPases, GTPases, and dUTPases, where they assist in the catalysis of the gamma phosphate or gamma and beta phosphates from ATP or GTP, which creates a release of energy which can be used to perform cellular work. They are also found in GTPase-activating proteins (GAP). Thus, they are essential for many forms of life, and are highly conserved. Arginine fingers function through non-covalent interactions. They may also assist in dimerization, and while they are found in a wide variety of enzymes, they are not ubiquitous.
Lynette Cegelski is an American physical chemist and chemical biologist who studies extracellular structures such as biofilms and membrane proteins. She is an associate professor of chemistry and, by courtesy, of chemical engineering at Stanford University. She is a Stanford Bio-X and Stanford ChEM-H affiliated faculty member.

Peter R. Buseck is a Regents Professor in the School of Molecular Sciences (SMS) at Arizona State University (ASU). He is a pioneering researcher in the application of transmission electron microscopy to mineralogy, meteoritics, fullerenes and atmospheric chemistry. In 2019 Buseck was awarded the Roebling Medal, the highest award of the Mineralogical Society of America. The scientific journal Nature recognized Buseck's 1978 paper as a milestone in crystallography.
Susanne Marie Rafelski is an American biochemist. Rafelski studied biochemistry at the University of Arizona with David Galbraith. She obtained her PhD in 2005 from Stanford University, under supervision of Julie Theriot.
Emma Lundberg is a Swedish cell biologist who is a professor at KTH Royal Institute of Technology and Director of Cell Profiling at the Science for Life Laboratory. Her research considers spatial proteomics and cell biology, making use of an antibody-based approach to assess fundamental aspects of human biology. She looks to understand why certain variations in human proteins can cause disease.
Song Lin is a Chinese-American organic electrochemist who is an associate professor at Cornell University. His research involves the development of new synthetic organic methodologies that utilize electrochemistry to forge new chemical bonds. He is an Associate Editor of the journal Organic Letters, and serves on the Early Career Advisory Board of Chemistry - A European Journal. He was named by Chemical & Engineering News as one of their Trailblazers of 2022, a feature highlighting LGBTQ+ chemists in academia.











