Phenylketonuria (PKU) is an inborn error of metabolism that results in decreased metabolism of the amino acid phenylalanine. Untreated PKU can lead to intellectual disability, seizures, behavioral problems, and mental disorders. It may also result in a musty smell and lighter skin. A baby born to a mother who has poorly treated PKU may have heart problems, a small head, and low birth weight.
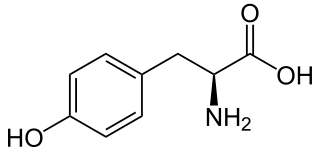
L-Tyrosine or tyrosine or 4-hydroxyphenylalanine is one of the 20 standard amino acids that are used by cells to synthesize proteins. It is a non-essential amino acid with a polar side group. The word "tyrosine" is from the Greek tyrós, meaning cheese, as it was first discovered in 1846 by German chemist Justus von Liebig in the protein casein from cheese. It is called tyrosyl when referred to as a functional group or side chain. While tyrosine is generally classified as a hydrophobic amino acid, it is more hydrophilic than phenylalanine. It is encoded by the codons UAC and UAU in messenger RNA.

Phenylalanine is an essential α-amino acid with the formula C
9H
11NO
2. It can be viewed as a benzyl group substituted for the methyl group of alanine, or a phenyl group in place of a terminal hydrogen of alanine. This essential amino acid is classified as neutral, and nonpolar because of the inert and hydrophobic nature of the benzyl side chain. The L-isomer is used to biochemically form proteins coded for by DNA. Phenylalanine is a precursor for tyrosine, the monoamine neurotransmitters dopamine, norepinephrine (noradrenaline), and epinephrine (adrenaline), and the skin pigment melanin. It is encoded by the codons UUU and UUC.
Phenylalanine hydroxylase. (PAH) (EC 1.14.16.1) is an enzyme that catalyzes the hydroxylation of the aromatic side-chain of phenylalanine to generate tyrosine. PAH is one of three members of the biopterin-dependent aromatic amino acid hydroxylases, a class of monooxygenase that uses tetrahydrobiopterin (BH4, a pteridine cofactor) and a non-heme iron for catalysis. During the reaction, molecular oxygen is heterolytically cleaved with sequential incorporation of one oxygen atom into BH4 and phenylalanine substrate. In humans, mutations in its encoding gene, PAH, can lead to the metabolic disorder phenylketonuria.

Dehydroalanine is a dehydroamino acid. It does not exist in its free form, but it occurs naturally as a residue found in peptides of microbial origin. As an amino acid residue, it is unusual because it has an unsaturated backbone.

Cinnamaldehyde is an organic compound with the formula() C6H5CH=CHCHO. Occurring naturally as predominantly the trans (E) isomer, it gives cinnamon its flavor and odor. It is a phenylpropanoid that is naturally synthesized by the shikimate pathway. This pale yellow, viscous liquid occurs in the bark of cinnamon trees and other species of the genus Cinnamomum. The essential oil of cinnamon bark is about 90% cinnamaldehyde. Cinnamaldehyde decomposes to styrene because of oxidation as a result of bad storage or transport conditions. Styrene especially forms in high humidity and high temperatures. This is the reason why cinnamon contains small amounts of styrene.
Shikimic acid, more commonly known as its anionic form shikimate, is a cyclohexene, a cyclitol and a cyclohexanecarboxylic acid. It is an important biochemical metabolite in plants and microorganisms. Its name comes from the Japanese flower shikimi, from which it was first isolated in 1885 by Johan Fredrik Eykman. The elucidation of its structure was made nearly 50 years later.

Apigenin (4′,5,7-trihydroxyflavone), found in many plants, is a natural product belonging to the flavone class that is the aglycone of several naturally occurring glycosides. It is a yellow crystalline solid that has been used to dye wool.

The phenylpropanoids are a diverse family of organic compounds that are synthesized by plants from the amino acids phenylalanine and tyrosine. Their name is derived from the six-carbon, aromatic phenyl group and the three-carbon propene tail of coumaric acid, which is the central intermediate in phenylpropanoid biosynthesis. From 4-coumaroyl-CoA emanates the biosynthesis of myriad natural products including lignols, flavonoids, isoflavonoids, coumarins, aurones, stilbenes, catechin, and phenylpropanoids. The coumaroyl component is produced from cinnamic acid.

An aromatic amino acid is an amino acid that includes an aromatic ring.

Histidine ammonia-lyase is an enzyme that in humans is encoded by the HAL gene. It converts histidine into ammonia and urocanic acid. Its systematic name is L-histidine ammonia-lyase (urocanate-forming).

Cystathionine beta-lyase, also commonly referred to as CBL or β-cystathionase, is an enzyme that primarily catalyzes the following α,β-elimination reaction

Threonine ammonia-lyase (EC 4.3.1.19, systematic name L-threonine ammonia-lyase (2-oxobutanoate-forming), also commonly referred to as threonine deaminase or threonine dehydratase, is an enzyme responsible for catalyzing the conversion of L-threonine into α-ketobutyrate and ammonia:

Arogenate dehydratase (ADT) (EC 4.2.1.91) is an enzyme that catalyzes the chemical reaction
In enzymology, glutamate-prephenate aminotransferase is an enzyme that catalyzes the chemical reaction

Rosavin are a family of cinnamyl mono- and diglycosides that are key ingredients of Rhodiola rosea L.,. R. rosea is an important medicinal plant commonly used throughout Europe, Asia, and North America, that has been recognized as a botanical adaptogen by the European Medicines Agency. Rosavin production is specific to R. rosea and R. sachalinenis, and the biosynthesis of these glycosides occurs spontaneously in Rhodiola roots and rhizomes. The production of rosavins increases in plants as they get older, and the amount of the cinnamyl alcohol glycosides depends on the place of origin of the plant.

Biopterin-dependent aromatic amino acid hydroxylases (AAAH) are a family of aromatic amino acid hydroxylase enzymes which includes phenylalanine 4-hydroxylase, tyrosine 3-hydroxylase, and tryptophan 5-hydroxylase. These enzymes primarily hydroxylate the amino acids L-phenylalanine, L-tyrosine, and L-tryptophan, respectively.
The biosynthesis of phenylpropanoids involves a number of enzymes.
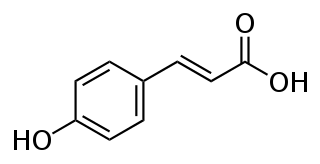
Tyrosine ammonia lyase (EC 4.3.1.23, L-tyrosine ammonia-lyase, TAL or Tyrase) is an enzyme in the natural phenols biosynthesis pathway. It transforms L-tyrosine into p-coumaric acid.
Phenylalanine/tyrosine ammonia-lyase (EC 4.3.1.25, PTAL, bifunctional PAL) is an enzyme with systematic name L-phenylalanine(or L-tyrosine):trans-cinnamate(or trans-p-hydroxycinnamate) ammonia-lyase. This enzyme catalyses the following chemical reaction