
In biochemistry, denaturation is a process in which proteins or nucleic acids lose folded structure present in their native state due to various factors, including application of some external stress or compound, such as a strong acid or base, a concentrated inorganic salt, an organic solvent, agitation and radiation, or heat. If proteins in a living cell are denatured, this results in disruption of cell activity and possibly cell death. Protein denaturation is also a consequence of cell death. Denatured proteins can exhibit a wide range of characteristics, from conformational change and loss of solubility or dissociation of cofactors to aggregation due to the exposure of hydrophobic groups. The loss of solubility as a result of denaturation is called coagulation. Denatured proteins lose their 3D structure, and therefore, cannot function.

In organic chemistry, a peptide bond is an amide type of covalent chemical bond linking two consecutive alpha-amino acids from C1 of one alpha-amino acid and N2 of another, along a peptide or protein chain.
In chemistry, a zwitterion, also called an inner salt or dipolar ion, is a molecule that contains an equal number of positively and negatively charged functional groups. With amino acids, for example, in solution a chemical equilibrium will be established between the "parent" molecule and the zwitterion.
Proline (symbol Pro or P) is an organic acid classed as a proteinogenic amino acid (used in the biosynthesis of proteins), although it does not contain the amino group -NH
2 but is rather a secondary amine. The secondary amine nitrogen is in the protonated form (NH2+) under biological conditions, while the carboxyl group is in the deprotonated −COO− form. The "side chain" from the α carbon connects to the nitrogen forming a pyrrolidine loop, classifying it as a aliphatic amino acid. It is non-essential in humans, meaning the body can synthesize it from the non-essential amino acid L-glutamate. It is encoded by all the codons starting with CC (CCU, CCC, CCA, and CCG).
In chemistry, a disulfide is a compound containing a R−S−S−R′ functional group or the S2−
2 anion. The linkage is also called an SS-bond or sometimes a disulfide bridge and usually derived from two thiol groups.

Flavins refers generally to the class of organic compounds containing the tricyclic heterocycle isoalloxazine or its isomer alloxazine, and derivatives thereof. The biochemical source of flavin is the yellow B vitamin riboflavin. The flavin moiety is often attached with an adenosine diphosphate to form flavin adenine dinucleotide (FAD), and, in other circumstances, is found as flavin mononucleotide, a phosphorylated form of riboflavin. It is in one or the other of these forms that flavin is present as a prosthetic group in flavoproteins. Despite the similar names, flavins are chemically and biologically distinct from the flavanoids, and the flavonols.

Rhodopsin, also known as visual purple, is a protein encoded by the RHO gene and a G-protein-coupled receptor (GPCR). It is the opsin of the rod cells in the retina and a light-sensitive receptor protein that triggers visual phototransduction in rods. Rhodopsin mediates dim light vision and thus is extremely sensitive to light. When rhodopsin is exposed to light, it immediately photobleaches. In humans, it is regenerated fully in about 30 minutes, after which the rods are more sensitive. Defects in the rhodopsin gene cause eye diseases such as retinitis pigmentosa and congenital stationary night blindness.

Photochemistry is the branch of chemistry concerned with the chemical effects of light. Generally, this term is used to describe a chemical reaction caused by absorption of ultraviolet, visible (400–750 nm), or infrared radiation (750–2500 nm).

Bacteriorhodopsin (Bop) is a protein used by Archaea, most notably by haloarchaea, a class of the Euryarchaeota. It acts as a proton pump; that is, it captures light energy and uses it to move protons across the membrane out of the cell. The resulting proton gradient is subsequently converted into chemical energy.
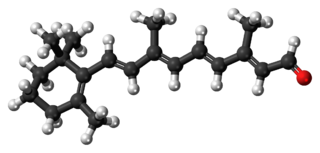
Retinal is a polyene chromophore. Retinal, bound to proteins called opsins, is the chemical basis of visual phototransduction, the light-detection stage of visual perception (vision).
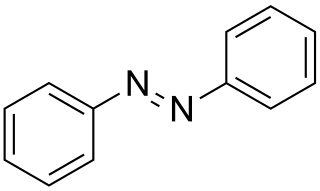
Azobenzene is a photoswitchable chemical compound composed of two phenyl rings linked by a N=N double bond. It is the simplest example of an aryl azo compound. The term 'azobenzene' or simply 'azo' is often used to refer to a wide class of similar compounds. These azo compounds are considered as derivatives of diazene (diimide), and are sometimes referred to as 'diazenes'. The diazenes absorb light strongly and are common dyes. Different classes of azo dyes exist, most notably the ones substituted with heteroaryl rings.

Aconitase is an enzyme that catalyses the stereo-specific isomerization of citrate to isocitrate via cis-aconitate in the tricarboxylic acid cycle, a non-redox-active process.
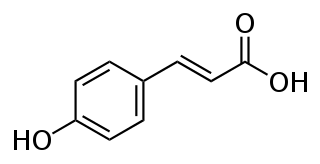
p-Coumaric acid is an organic compound with the formula HOC6H4CH=CHCO2H. It is one of the three isomers of hydroxycinnamic acid. It is a white solid that is only slightly soluble in water but very soluble in ethanol and diethyl ether.
In chemistry, persulfide refers to the functional group R-S-S-H. Persulfides are intermediates in the biosynthesis of iron-sulfur proteins and are invoked as precursors to hydrogen sulfide, a signaling molecule.
Retinylidene proteins, or rhodopsins in a broad sense, are proteins that use retinal as a chromophore for light reception. They are the molecular basis for a variety of light-sensing systems from phototaxis in flagellates to eyesight in animals. Retinylidene proteins include all forms of opsin and rhodopsin. While rhodopsin in the narrow sense refers to a dim-light visual pigment found in vertebrates, usually on rod cells, rhodopsin in the broad sense refers to any molecule consisting of an opsin and a retinal chromophore in the ground state. When activated by light, the chromophore is isomerized, at which point the molecule as a whole is no longer rhodopsin, but a related molecule such as metarhodopsin. However, it remains a retinylidene protein. The chromophore then separates from the opsin, at which point the bare opsin is a retinylidene protein. Thus, the molecule remains a retinylidene protein throughout the phototransduction cycle.

In enzymology, a steroid Δ5-isomerase is an enzyme that catalyzes the chemical reaction

Cystathionine beta-lyase, also commonly referred to as CBL or β-cystathionase, is an enzyme that primarily catalyzes the following α,β-elimination reaction

RPE-retinal G protein-coupled receptor also known as RGR-opsin is a protein that in humans is encoded by the RGR gene. RGR-opsin is a member of the rhodopsin-like receptor subfamily of GPCR. Like other opsins which bind retinaldehyde, it contains a conserved lysine residue in the seventh transmembrane domain. RGR-opsin comes in different isoforms produced by alternative splicing.
Halorhodospira halophila is a species of Halorhodospira distinguished by its ability to grow optimally in an environment of 15–20% salinity. It was formerly called Ectothiorhodospira halophila. It is an anaerobic, rod-shaped Gram-negative bacterium. H. halophila has a flagellum.

Archaerhodopsin proteins are a family of retinal-containing photoreceptors found in the archaea genera Halobacterium and Halorubrum. Like the homologous bacteriorhodopsin (bR) protein, archaerhodopsins harvest energy from sunlight to pump H+ ions out of the cell, establishing a proton motive force that is used for ATP synthesis. They have some structural similarities to the mammalian G protein-coupled receptor protein rhodopsin, but are not true homologs.














