
The cell is the basic structural and functional unit of all forms of life. Every cell consists of cytoplasm enclosed within a membrane; many cells contain organelles, each with a specific function. The term comes from the Latin word cellula meaning 'small room'. Most cells are only visible under a microscope. Cells emerged on Earth about 4 billion years ago. All cells are capable of replication, protein synthesis, and motility.
In cell biology, an organelle is a specialized subunit, usually within a cell, that has a specific function. The name organelle comes from the idea that these structures are parts of cells, as organs are to the body, hence organelle, the suffix -elle being a diminutive. Organelles are either separately enclosed within their own lipid bilayers or are spatially distinct functional units without a surrounding lipid bilayer. Although most organelles are functional units within cells, some function units that extend outside of cells are often termed organelles, such as cilia, the flagellum and archaellum, and the trichocyst.

A flagellum is a hairlike appendage that protrudes from certain plant and animal sperm cells, from fungal spores (zoospores), and from a wide range of microorganisms to provide motility. Many protists with flagella are known as flagellates.
The evolution of flagella is of great interest to biologists because the three known varieties of flagella – each represent a sophisticated cellular structure that requires the interaction of many different systems.

ATPases (EC 3.6.1.3, Adenosine 5'-TriPhosphatase, adenylpyrophosphatase, ATP monophosphatase, triphosphatase, SV40 T-antigen, ATP hydrolase, complex V (mitochondrial electron transport), (Ca2+ + Mg2+)-ATPase, HCO3−-ATPase, adenosine triphosphatase) are a class of enzymes that catalyze the decomposition of ATP into ADP and a free phosphate ion or the inverse reaction. This dephosphorylation reaction releases energy, which the enzyme (in most cases) harnesses to drive other chemical reactions that would not otherwise occur. This process is widely used in all known forms of life.

The cytoskeleton is a complex, dynamic network of interlinking protein filaments present in the cytoplasm of all cells, including those of bacteria and archaea. In eukaryotes, it extends from the cell nucleus to the cell membrane and is composed of similar proteins in the various organisms. It is composed of three main components: microfilaments, intermediate filaments, and microtubules, and these are all capable of rapid growth or disassembly depending on the cell's requirements.

ATP synthase is an enzyme that catalyzes the formation of the energy storage molecule adenosine triphosphate (ATP) using adenosine diphosphate (ADP) and inorganic phosphate (Pi). ATP synthase is a molecular machine. The overall reaction catalyzed by ATP synthase is:
The periplasm is a concentrated gel-like matrix in the space between the inner cytoplasmic membrane and the bacterial outer membrane called the periplasmic space in gram-negative bacteria. Using cryo-electron microscopy it has been found that a much smaller periplasmic space is also present in gram-positive bacteria, between cell wall and the plasma membrane. The periplasm may constitute up to 40% of the total cell volume of gram-negative bacteria, but is a much smaller percentage in gram-positive bacteria.
Members of the genus Selenomonas are referred to trivially as selenomonads. The genus Selenomonas constitutes a group of motile crescent-shaped bacteria and includes species living in the gastrointestinal tracts of animals, in particular the ruminants. A number of smaller forms discovered with the light microscope are now in culture but many, especially the large selenomonads are not, owing to their fastidious and incompletely known growth requirements.
A bacterium, despite its simplicity, contains a well-developed cell structure which is responsible for some of its unique biological structures and pathogenicity. Many structural features are unique to bacteria and are not found among archaea or eukaryotes. Because of the simplicity of bacteria relative to larger organisms and the ease with which they can be manipulated experimentally, the cell structure of bacteria has been well studied, revealing many biochemical principles that have been subsequently applied to other organisms.

Vacuolar-type ATPase (V-ATPase) is a highly conserved evolutionarily ancient enzyme with remarkably diverse functions in eukaryotic organisms. V-ATPases acidify a wide array of intracellular organelles and pumps protons across the plasma membranes of numerous cell types. V-ATPases couple the energy of ATP hydrolysis to proton transport across intracellular and plasma membranes of eukaryotic cells. It is generally seen as the polar opposite of ATP synthase because ATP synthase is a proton channel that uses the energy from a proton gradient to produce ATP. V-ATPase however, is a proton pump that uses the energy from ATP hydrolysis to produce a proton gradient.
An undulipodium or undulopodium, or a 9+2 organelle is a motile filamentous extracellular projection of eukaryotic cells. It is basically synonymous to flagella and cilia which are differing terms for similar molecular structures used on different types of cells, and usually correspond to different waveforms.
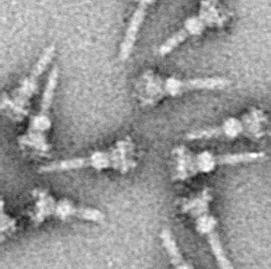
The type III secretion system is one of the bacterial secretion systems used by bacteria to secrete their effector proteins into the host's cells to promote virulence and colonisation. While the type III secretion system has been widely regarded as equivalent to the injectisome, many argue that the injectisome is only part of the type III secretion system, which also include structures like the flagellar export apparatus. The T3SS is a needle-like protein complex found in several species of pathogenic gram-negative bacteria.

The alpha and beta subunits are found in the F1, V1, and A1 complexes of F-, V- and A-ATPases, respectively, as well as flagellar (T3SS) ATPase and the termination factor Rho. The subunits make up a ring that contains the ATP-hydrolyzing catalytic core. The F-ATPases, V-ATPases and A-ATPases are composed of two linked complexes: the F1, V1 or A1 complex containsthat synthesizes/hydrolyses ATP, and the Fo, Vo or Ao complex that forms the membrane-spanning pore. The F-, V- and A-ATPases all contain rotary motors, one that drives proton translocation across the membrane and one that drives ATP synthesis/hydrolysis.

Bacterial motility is the ability of bacteria to move independently using metabolic energy. Most motility mechanisms that evolved among bacteria also evolved in parallel among the archaea. Most rod-shaped bacteria can move using their own power, which allows colonization of new environments and discovery of new resources for survival. Bacterial movement depends not only on the characteristics of the medium, but also on the use of different appendages to propel. Swarming and swimming movements are both powered by rotating flagella. Whereas swarming is a multicellular 2D movement over a surface and requires the presence of surfactants, swimming is movement of individual cells in liquid environments.

A prokaryote is a single-cell organism whose cell lacks a nucleus and other membrane-bound organelles. The word prokaryote comes from the Ancient Greek πρό 'before' and κάρυον 'nut, kernel'. In the two-empire system arising from the work of Édouard Chatton, prokaryotes were classified within the empire Prokaryota. But in the three-domain system, based upon molecular analysis, prokaryotes are divided into two domains: Bacteria and Archaea. Organisms with nuclei are placed in a third domain, Eukaryota.

Several organisms are capable of rolling locomotion. However, true wheels and propellers—despite their utility in human vehicles—do not play a significant role in the movement of living things. Biologists have offered several explanations for the apparent absence of biological wheels, and wheeled creatures have appeared often in speculative fiction.
The archaellum is a unique structure on the cell surface of many archaea that allows for swimming motility. The archaellum consists of a rigid helical filament that is attached to the cell membrane by a molecular motor. This molecular motor – composed of cytosolic, membrane, and pseudo-periplasmic proteins – is responsible for the assembly of the filament and, once assembled, for its rotation. The rotation of the filament propels archaeal cells in liquid medium, in a manner similar to the propeller of a boat. The bacterial analog of the archaellum is the flagellum, which is also responsible for their swimming motility and can also be compared to a rotating corkscrew. Although the movement of archaella and flagella is sometimes described as "whip-like", this is incorrect, as only cilia from Eukaryotes move in this manner. Indeed, even "flagellum" is a misnomer, as bacterial flagella also work as propeller-like structures.

Bacterial secretion systems are protein complexes present on the cell membranes of bacteria for secretion of substances. Specifically, they are the cellular devices used by pathogenic bacteria to secrete their virulence factors to invade the host cells. They can be classified into different types based on their specific structure, composition and activity. Generally, proteins can be secreted through two different processes. One process is a one-step mechanism in which proteins from the cytoplasm of bacteria are transported and delivered directly through the cell membrane into the host cell. Another involves a two-step activity in which the proteins are first transported out of the inner cell membrane, then deposited in the periplasm, and finally through the outer cell membrane into the host cell.
Monocercomonas is a Parabasalian genus belonging to the order Trichomonadida. It presents four flagella, three forward-facing and one trailing, without the presence of a costa or any kind of undulating membrane. Monocercomonas is found in animal guts. and is susceptible to cause Monocercomoniasis in reptiles












