Related Research Articles
An allele is one of two, or more, forms of a given gene variant. For example, the ABO blood grouping is controlled by the ABO gene, which has six common alleles. Nearly every living human's phenotype for the ABO gene is some combination of just these six alleles. An allele is one of two, or more, versions of the same gene at the same place on a chromosome. It can also refer to different sequence variations for several-hundred base-pair or more region of the genome that codes for a protein. Alleles can come in different extremes of size. At the lowest possible size an allele can be a single nucleotide polymorphism (SNP). At the higher end, it can be up to several thousand base-pairs long. Most alleles result in little or no observable change in the function of the protein the gene codes for.
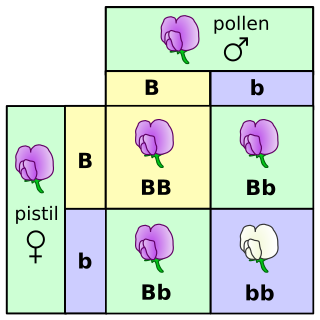
The genotype of an organism is its complete set of genetic material. However, the term is often used to refer to a single gene or set of genes, such as the genotype for eye color. The genes partly determine the observable characteristics of an organism, such as hair color, height, etc. An example of a characteristic determined by a genotype is the petal color in a pea plant. The collection of all genetic possibilities for a single trait are called alleles; two alleles for petal color are purple and white.

In biology, a mutation is an alteration in the nucleotide sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mitosis, or meiosis or other types of damage to DNA, which then may undergo error-prone repair, cause an error during other forms of repair, or cause an error during replication. Mutations may also result from insertion or deletion of segments of DNA due to mobile genetic elements.

A phenotypic trait, simply trait, or character state is a distinct variant of a phenotypic characteristic of an organism; it may be either inherited or determined environmentally, but typically occurs as a combination of the two. For example, eye color is a character of an organism, while blue, brown and hazel are traits.
Heterochromatin is a tightly packed form of DNA or condensed DNA, which comes in multiple varieties. These varieties lie on a continuum between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a role in the expression of genes. Because it is tightly packed, it was thought to be inaccessible to polymerases and therefore not transcribed, however according to Volpe et al. (2002), and many other papers since, much of this DNA is in fact transcribed, but it is continuously turned over via RNA-induced transcriptional silencing (RITS). Recent studies with electron microscopy and OsO4 staining reveal that the dense packing is not due to the chromatin.

In genetics, dominance is the phenomenon of one variant (allele) of a gene on a chromosome masking or overriding the effect of a different variant of the same gene on the other copy of the chromosome. The first variant is termed dominant and the second recessive. This state of having two different variants of the same gene on each chromosome is originally caused by a mutation in one of the genes, either new or inherited. The terms autosomal dominant or autosomal recessive are used to describe gene variants on non-sex chromosomes (autosomes) and their associated traits, while those on sex chromosomes (allosomes) are termed X-linked dominant, X-linked recessive or Y-linked; these have an inheritance and presentation pattern that depends on the sex of both the parent and the child. Since there is only one copy of the Y chromosome, Y-linked traits cannot be dominant nor recessive. Additionally, there are other forms of dominance such as incomplete dominance, in which a gene variant has a partial effect compared to when it is present on both chromosomes, and co-dominance, in which different variants on each chromosome both show their associated traits.
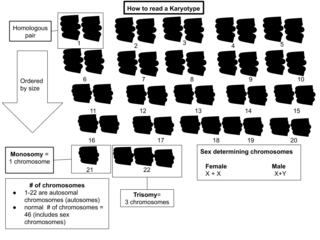
A karyotype is a preparation of the complete set of metaphase chromosomes in the cells of a species or in an individual organism, sorted by length, centromere location and other features. and for a test that detects this complement or counts the number of chromosomes. Karyotyping is the process by which a karyotype is prepared from photographs of chromosomes, in order to determine the chromosome complement of an individual, including the number of chromosomes and any abnormalities.
A genetic screen or mutagenesis screen is an experimental technique used to identify and select for individuals who possess a phenotype of interest in a mutagenized population. Hence a genetic screen is a type of phenotypic screen. Genetic screens can provide important information on gene function as well as the molecular events that underlie a biological process or pathway. While genome projects have identified an extensive inventory of genes in many different organisms, genetic screens can provide valuable insight as to how those genes function.

Non-Mendelian inheritance is any pattern of inheritance in which traits do not segregate in accordance with Mendel's laws. These laws describe the inheritance of traits linked to single genes on chromosomes in the nucleus. In Mendelian inheritance, each parent contributes one of two possible alleles for a trait. If the genotypes of both parents in a genetic cross are known, Mendel's laws can be used to determine the distribution of phenotypes expected for the population of offspring. There are several situations in which the proportions of phenotypes observed in the progeny do not match the predicted values.
Transvection is an epigenetic phenomenon that results from an interaction between an allele on one chromosome and the corresponding allele on the homologous chromosome. Transvection can lead to either gene activation or repression. It can also occur between nonallelic regions of the genome as well as regions of the genome that are not transcribed.

In genetics, a locus is a specific, fixed position on a chromosome where a particular gene or genetic marker is located. Each chromosome carries many genes, with each gene occupying a different position or locus; in humans, the total number of protein-coding genes in a complete haploid set of 23 chromosomes is estimated at 19,000–20,000.
P elements are transposable elements that were discovered in Drosophila as the causative agents of genetic traits called hybrid dysgenesis. The transposon is responsible for the P trait of the P element and it is found only in wild flies. They are also found in many other eukaryotes.
A transgene is a gene that has been transferred naturally, or by any of a number of genetic engineering techniques from one organism to another. The introduction of a transgene, in a process known as transgenesis, has the potential to change the phenotype of an organism. Transgene describes a segment of DNA containing a gene sequence that has been isolated from one organism and is introduced into a different organism. This non-native segment of DNA may either retain the ability to produce RNA or protein in the transgenic organism or alter the normal function of the transgenic organism's genetic code. In general, the DNA is incorporated into the organism's germ line. For example, in higher vertebrates this can be accomplished by injecting the foreign DNA into the nucleus of a fertilized ovum. This technique is routinely used to introduce human disease genes or other genes of interest into strains of laboratory mice to study the function or pathology involved with that particular gene.
An insulator is a type of cis-regulatory element known as a long-range regulatory element. Found in multicellular eukaryotes and working over distances from the promoter element of the target gene, an insulator is typically 300 bp to 2000 bp in length. Insulators contain clustered binding sites for sequence specific DNA-binding proteins and mediate intra- and inter-chromosomal interactions.
Position-effect variegation (PEV) is a variegation caused by the silencing of a gene in some cells through its abnormal juxtaposition with heterochromatin via rearrangement or transposition. It is also associated with changes in chromatin conformation.
This glossary of genetics is a list of definitions of terms and concepts commonly used in the study of genetics and related disciplines in biology, including molecular biology and evolutionary biology. It is intended as introductory material for novices; for more specific and technical detail, see the article corresponding to each term. For related terms, see Glossary of evolutionary biology.

The GAL4-UAS system is a biochemical method used to study gene expression and function in organisms such as the fruit fly. It has also been adapted to study receptor chemical-binding functions in vitro in cell culture. It was developed by Hitoshi Kakidani and Mark Ptashne, and Nicholas Webster and Pierre Chambon in 1988, then adapted by Andrea Brand and Norbert Perrimon in 1993 and is considered a powerful technique for studying the expression of genes. The system has two parts: the Gal4 gene, encoding the yeast transcription activator protein Gal4, and the UAS, an enhancer to which GAL4 specifically binds to activate gene transcription.

Brainbow is a process by which individual neurons in the brain can be distinguished from neighboring neurons using fluorescent proteins. By randomly expressing different ratios of red, green, and blue derivatives of green fluorescent protein in individual neurons, it is possible to flag each neuron with a distinctive color. This process has been a major contribution to the field of neural connectomics.
Transposons are semi-parasitic DNA sequences which can replicate and spread through the host's genome. They can be harnessed as a genetic tool for analysis of gene and protein function. The use of transposons is well-developed in Drosophila and in Thale cress and bacteria such as Escherichia coli.
Q-system is a genetic tool that allows to express transgenes in a living organism. Originally the Q-system was developed for use in the vinegar fly Drosophila melanogaster, and was rapidly adapted for use in cultured mammalian cells, zebrafish, worms and mosquitoes. The Q-system utilizes genes from the qa cluster of the bread fungus Neurospora crassa, and consists of four components: the transcriptional activator (QF/QF2/QF2w), the enhancer QUAS, the repressor QS, and the chemical de-repressor quinic acid. Similarly to GAL4/UAS and LexA/LexAop, the Q-system is a binary expression system that allows to express reporters or effectors in a defined subpopulation of cells with the purpose of visualising these cells or altering their function. In addition, GAL4/UAS, LexA/LexAop and the Q-system function independently of each other and can be used simultaneously to achieve a desired pattern of reporter expression, or to express several reporters in different subsets of cells.
References
- ↑ Weiler K, Wakimoto B (1995). "Heterochromatin and gene expression in Drosophila". Annu Rev Genet. 29 (1): 577–605. doi:10.1146/annurev.ge.29.120195.003045. PMID 8825487.