Related Research Articles

A bacteriophage, also known informally as a phage, is a virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν, meaning "to devour". Bacteriophages are composed of proteins that encapsulate a DNA or RNA genome, and may have structures that are either simple or elaborate. Their genomes may encode as few as four genes and as many as hundreds of genes. Phages replicate within the bacterium following the injection of their genome into its cytoplasm.

A capsid is the protein shell of a virus, enclosing its genetic material. It consists of several oligomeric (repeating) structural subunits made of protein called protomers. The observable 3-dimensional morphological subunits, which may or may not correspond to individual proteins, are called capsomeres. The proteins making up the capsid are called capsid proteins or viral coat proteins (VCP). The capsid and inner genome is called the nucleocapsid.

Chaperone proteins participate in the folding of over half of all mammalian proteins. In molecular biology, molecular chaperones are proteins that assist the conformational folding or unfolding and the assembly or disassembly of other macromolecular structures. Chaperones are present when the macromolecules perform their normal biological functions and have correctly completed the processes of folding and/or assembly. The chaperones are concerned primarily with protein folding. The first protein to be called a chaperone assists the assembly of nucleosomes from folded histones and DNA and such assembly chaperones, especially in the nucleus, are concerned with the assembly of folded subunits into oligomeric structures.
A cosmid is a type of hybrid plasmid that contains a Lambda phage cos sequence. They are often used as a cloning vector in genetic engineering. Cosmids can be used to build genomic libraries. They were first described by Collins and Hohn in 1978. Cosmids can contain 37 to 52 kb of DNA, limits based on the normal bacteriophage packaging size. They can replicate as plasmids if they have a suitable origin of replication (ori): for example SV40 ori in mammalian cells, ColE1 ori for double-stranded DNA replication, or f1 ori for single-stranded DNA replication in prokaryotes. They frequently also contain a gene for selection such as antibiotic resistance, so that the transformed cells can be identified by plating on a medium containing the antibiotic. Those cells which did not take up the cosmid would be unable to grow.

Escherichia virus T4 is a species of bacteriophages that infect Escherichia coli bacteria. It is a double-stranded DNA virus in the subfamily Tevenvirinae from the family Myoviridae. T4 is capable of undergoing only a lytic lifecycle and not the lysogenic lifecycle. The species was formerly named T-even bacteriophage, a name which also encompasses, among other strains, Enterobacteria phage T2, Enterobacteria phage T4 and Enterobacteria phage T6.

A peptidomimetic is a small protein-like chain designed to mimic a peptide. They typically arise either from modification of an existing peptide, or by designing similar systems that mimic peptides, such as peptoids and β-peptides. Irrespective of the approach, the altered chemical structure is designed to advantageously adjust the molecular properties such as stability or biological activity. This can have a role in the development of drug-like compounds from existing peptides. These modifications involve changes to the peptide that will not occur naturally. Based on their similarity with the precursor peptide, peptidomimetics can be grouped into four classes where A features the most and D the least similarities. Classes A and B involve peptide-like scaffolds, while classes C and D include small molecules.

Phage display is a laboratory technique for the study of protein–protein, protein–peptide, and protein–DNA interactions that uses bacteriophages to connect proteins with the genetic information that encodes them. In this technique, a gene encoding a protein of interest is inserted into a phage coat protein gene, causing the phage to "display" the protein on its outside while containing the gene for the protein on its inside, resulting in a connection between genotype and phenotype. These displaying phages can then be screened against other proteins, peptides or DNA sequences, in order to detect interaction between the displayed protein and those other molecules. In this way, large libraries of proteins can be screened and amplified in a process called in vitro selection, which is analogous to natural selection.

Filamentous bacteriophage is a family of viruses (Inoviridae) that infect bacteria. The phages are named for their filamentous shape, a worm-like chain, about 6 nm in diameter and about 1000-2000 nm long. The coat of the virion comprises five types of viral protein, which are located during phage assembly in the inner membrane of the host bacteria, and are added to the nascent virion as it extrudes through the membrane. The simplicity of this family makes it an attractive model system to study fundamental aspects of molecular biology, and it has also proven useful as a tool in immunology and nanotechnology.

The phi X 174 bacteriophage is a single-stranded DNA (ssDNA) virus that infects Escherichia coli, and the first DNA-based genome to be sequenced. This work was completed by Fred Sanger and his team in 1977. In 1962, Walter Fiers and Robert Sinsheimer had already demonstrated the physical, covalently closed circularity of ΦX174 DNA. Nobel prize winner Arthur Kornberg used ΦX174 as a model to first prove that DNA synthesized in a test tube by purified enzymes could produce all the features of a natural virus, ushering in the age of synthetic biology. In 1972–1974, Jerard Hurwitz, Sue Wickner, and Reed Wickner with collaborators identified the genes required to produce the enzymes to catalyze conversion of the single stranded form of the virus to the double stranded replicative form. In 2003, it was reported by Craig Venter's group that the genome of ΦX174 was the first to be completely assembled in vitro from synthesized oligonucleotides. The ΦX174 virus particle has also been successfully assembled in vitro. In 2012, it was shown how its highly overlapping genome can be fully decompressed and still remain functional.
Spiegelman's Monster is the name given to an RNA chain of only 218 nucleotides that is able to be reproduced by the RNA replication enzyme RNA-dependent RNA polymerase, also called RNA replicase. It is named after its creator, Sol Spiegelman, of the University of Illinois at Urbana-Champaign who first described it in 1965.
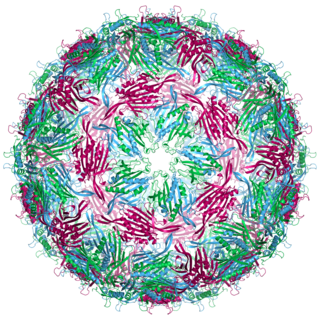
Bacteriophage MS2, commonly called MS2, is an icosahedral, positive-sense single-stranded RNA virus that infects the bacterium Escherichia coli and other members of the Enterobacteriaceae. MS2 is a member of a family of closely related bacterial viruses that includes bacteriophage f2, bacteriophage Qβ, R17, and GA.
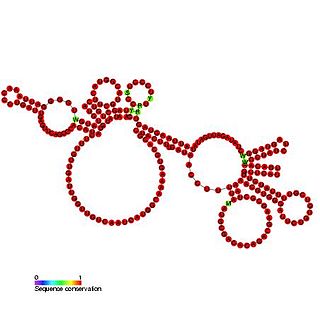
Bacteriophage pRNA is a ncRNA element. During replication of linear dsDNA viruses, the viral genome is packaged into the pre-formed viral procapsid. The packaging of DNA into the procapsid requires a molecular motor, which uses ATP as energy to accomplish the energetically unfavorable motion. In some bacteriophage, an RNA (pRNA) molecule is a vital component of this motor. Structural analyses of the packaging motor have demonstrated that the pRNA molecule has fivefold symmetry when attached to the prohead. The pRNA is thought to be bound by the capsid connector protein. Only the first 120 bases of the pRNA are essential for packing the viral DNA. The pRNA is proposed to be composed of two domains, one corresponding to the first 120 bases and the second to the remaining 50 bases. Nuclear cleavage occurs in the single strand region linking these two domains.
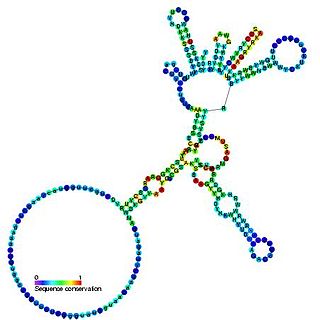
Group I introns are large self-splicing ribozymes. They catalyze their own excision from mRNA, tRNA and rRNA precursors in a wide range of organisms. The core secondary structure consists of nine paired regions (P1-P9). These fold to essentially two domains – the P4-P6 domain and the P3-P9 domain. The secondary structure mark-up for this family represents only this conserved core. Group I introns often have long open reading frames inserted in loop regions.

Bacteriophage Qbeta, commonly referred to as Qbeta or Qβ, is a positive-strand RNA virus which infects bacteria that have F-pili, most commonly Escherichia coli. Its linear genome is packaged into an icosahedral capsid with a diameter of 28 nm. Bacteriophage Qβ enters its host cell after binding to the side of the F-pilus.
SaPIs are a family of ~15 kb mobile genetic elements resident in the genomes of the vast majority of S. aureus strains. Much like bacteriophages, SaPIs can be transferred to uninfected cells and integrate into the host chromosome. Unlike the bacterial viruses, however, integrated SaPIs are mobilized by host infection with "helper" bacteriophages. SaPIs are used by the host bacteria to co-opt the phage reproduction cycle for their own genetic transduction and also inhibit phage reproduction in the process.
Enterobacteria phage P4 is a temperate bacteriophage strain of species Escherichia virus P2 within genus Peduovirus, subfamily Peduovirinae, family Myoviridae. It is a satellite virus, requiring P2-related helper phage to grow lytically.
Bacteriophage Mu, also known as mu phage or mu bacteriophage, is a muvirus of the family Myoviridae which has been shown to cause genetic transposition. It is of particular importance as its discovery in Escherichia coli by Larry Taylor was among the first observations of insertion elements in a genome. This discovery opened up the world to an investigation of transposable elements and their effects on a wide variety of organisms. While Mu was specifically involved in several distinct areas of research, the wider implications of transposition and insertion transformed the entire field of genetics.
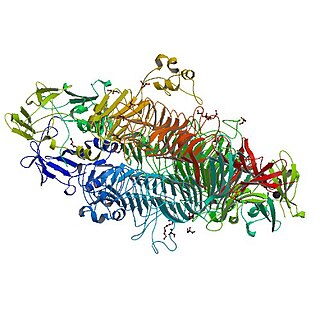
The tailspike protein (P22TSP) of Enterobacteria phage P22 mediates the recognition and adhesion between the bacteriophage and the surface of Salmonella enterica cells. It is anchored within the viral coat and recognizes the O-antigen portion of the lipopolysaccharide (LPS) on the outer-membrane of Gram-negative bacteria. It possesses endoglycanase activity, serving to shorten the length of the O-antigen during infection.

Helios Murialdo is a Chilean-Canadian molecular biologist, fiction writer, and ecologist. His research in the field of the assembly and structure of bacterial viruses contributed to the development of the first system for the cloning of human genes. He has published six novels and is a member of a group of conservationists that established a Natural Reserve in the central part of the Chilean Biodiversity Hotspot. Son of an Italian immigrant father and a Chilean mother of French descent, he has a brother and a sister. He is a member of the board of directors of the non-profit Fundación Ciencia & Vida, a scientific and technological institution with headquarters in Santiago, Chile. He is president of the NGO Corporación Altos de Cantillana, which manages the 26,000 acres of the Altos de Cantillana Natural Reserve in the coastal mountains of central Chile. He makes his home in this Natural Reserve, and spends the rest of the year in Toronto, Canada.
Charles Clifton Richardson is an American biochemist and professor at Harvard University. Richardson received his undergraduate education at Duke University, where he majored in medicine. He received his M.D. at Duke Medical School in 1960. Richardson works as a professor at Harvard Medical School, and he served as editor/associate editor of the Annual Review of Biochemistry from 1972 to 2003. Richardson received the American Chemical Society Award in Biological Chemistry in 1968, as well as numerous other accolades.
References
- ↑ Catalano, C. E.; Cue, D.; Feiss, M. (1995). "Virus DNA packaging: the strategy used by phage λ". Molecular Microbiology. 16 (6): 1075–1086. doi: 10.1111/j.1365-2958.1995.tb02333.x . PMID 8577244.
- ↑ Murialdo, H.; Becker, A. (1977). "Assembly of Biologically Active Proheads of Bacteriophage Lambda in vitro". Proceedings of the National Academy of Sciences. 74 (3): 906–910. Bibcode:1977PNAS...74..906M. doi: 10.1073/pnas.74.3.906 . PMC 430525 . PMID 265585.
- ↑ Ivanovska, I. L.; et al. (2004). "Bacteriophage capsids: Tough nanoshells with complex elastic properties". Proceedings of the National Academy of Sciences. 101 (20): 7600–7605. Bibcode:2004PNAS..101.7600I. doi: 10.1073/pnas.0308198101 . PMC 419652 . PMID 15133147.