Related Research Articles

The proteome is the entire set of proteins that is, or can be, expressed by a genome, cell, tissue, or organism at a certain time. It is the set of expressed proteins in a given type of cell or organism, at a given time, under defined conditions. Proteomics is the study of the proteome.

The human genome is a complete set of nucleic acid sequences for humans, encoded as DNA within the 23 chromosome pairs in cell nuclei and in a small DNA molecule found within individual mitochondria. These are usually treated separately as the nuclear genome and the mitochondrial genome. Human genomes include both protein-coding DNA genes and noncoding DNA. Haploid human genomes, which are contained in germ cells consist of three billion DNA base pairs, while diploid genomes have twice the DNA content. While there are significant differences among the genomes of human individuals, these are considerably smaller than the differences between humans and their closest living relatives, the bonobos and chimpanzees.
In bioinformatics, sequence analysis is the process of subjecting a DNA, RNA or peptide sequence to any of a wide range of analytical methods to understand its features, function, structure, or evolution. Methodologies used include sequence alignment, searches against biological databases, and others.

A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene or gene family and are the result of genetic differences. While many perform the same or similar biological roles, some isoforms have unique functions. A set of protein isoforms may be formed from alternative splicings, variable promoter usage, or other post-transcriptional modifications of a single gene; post-translational modifications are generally not considered. Through RNA splicing mechanisms, mRNA has the ability to select different protein-coding segments (exons) of a gene, or even different parts of exons from RNA to form different mRNA sequences. Each unique sequence produces a specific form of a protein.

In genetics, a single-nucleotide polymorphism is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently large fraction of the population, many publications do not apply such a frequency threshold.

Functional genomics is a field of molecular biology that attempts to describe gene functions and interactions. Functional genomics make use of the vast data generated by genomic and transcriptomic projects. Functional genomics focuses on the dynamic aspects such as gene transcription, translation, regulation of gene expression and protein–protein interactions, as opposed to the static aspects of the genomic information such as DNA sequence or structures. A key characteristic of functional genomics studies is their genome-wide approach to these questions, generally involving high-throughput methods rather than a more traditional "gene-by-gene" approach.
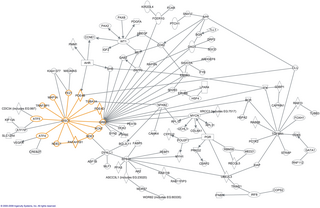
In molecular biology, an interactome is the whole set of molecular interactions in a particular cell. The term specifically refers to physical interactions among molecules but can also describe sets of indirect interactions among genes.
Protein subcellular localization prediction involves the prediction of where a protein resides in a cell, its subcellular localization.

Personalized medicine, also referred to as precision medicine, is a medical model that separates people into different groups—with medical decisions, practices, interventions and/or products being tailored to the individual patient based on their predicted response or risk of disease. The terms personalized medicine, precision medicine, stratified medicine and P4 medicine are used interchangeably to describe this concept though some authors and organisations use these expressions separately to indicate particular nuances.

Amos Bairoch is a Swiss bioinformatician and Professor of Bioinformatics at the Department of Human Protein Sciences of the University of Geneva where he leads the CALIPHO group at the Swiss Institute of Bioinformatics (SIB) combining bioinformatics, curation, and experimental efforts to functionally characterize human proteins.

Top-down proteomics is a method of protein identification that either uses an ion trapping mass spectrometer to store an isolated protein ion for mass measurement and tandem mass spectrometry (MS/MS) analysis or other protein purification methods such as two-dimensional gel electrophoresis in conjunction with MS/MS. Top-down proteomics is capable of identifying and quantitating unique proteoforms through the analysis of intact proteins. The name is derived from the similar approach to DNA sequencing. During mass spectrometry intact proteins are typically ionized by electrospray ionization and trapped in a Fourier transform ion cyclotron resonance, quadrupole ion trap or Orbitrap mass spectrometer. Fragmentation for tandem mass spectrometry is accomplished by electron-capture dissociation or electron-transfer dissociation. Effective fractionation is critical for sample handling before mass-spectrometry-based proteomics. Proteome analysis routinely involves digesting intact proteins followed by inferred protein identification using mass spectrometry (MS). Top-down MS (non-gel) proteomics interrogates protein structure through measurement of an intact mass followed by direct ion dissociation in the gas phase.

Claudin-8 is a protein that in humans is encoded by the CLDN8 gene. It belongs to the group of claudins.

DnaJ homolog subfamily B member 11 is a protein that in humans is encoded by the DNAJB11 gene.

Nucleotide exchange factor SIL1 is a protein that in humans is encoded by the SIL1 gene.
Edward Marcotte is a professor of biochemistry at The University of Texas at Austin, working in genetics, proteomics, and bioinformatics. Marcotte is an example of a computational biologist who also relies on experiments to validate bioinformatics-based predictions.
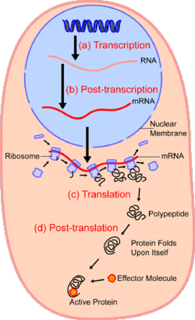
In the field of cellular biology, single-cell analysis is the study of genomics, transcriptomics, proteomics, metabolomics and cell–cell interactions at the single cell level. Due to the heterogeneity seen in both eukaryotic and prokaryotic cell populations, analyzing a single cell makes it possible to discover mechanisms not seen when studying a bulk population of cells. Technologies such as fluorescence-activated cell sorting (FACS) allow the precise isolation of selected single cells from complex samples, while high throughput single cell partitioning technologies, enable the simultaneous molecular analysis of hundreds or thousands of single unsorted cells; this is particularly useful for the analysis of transcriptome variation in genotypically identical cells, allowing the definition of otherwise undetectable cell subtypes. The development of new technologies is increasing our ability to analyze the genome and transcriptome of single cells, as well as to quantify their proteome and metabolome. Mass spectrometry techniques have become important analytical tools for proteomic and metabolomic analysis of single cells. Recent advances have enabled quantifying thousands of protein across hundreds of single cells, and thus make possible new types of analysis. In situ sequencing and fluorescence in situ hybridization (FISH) do not require that cells be isolated and are increasingly being used for analysis of tissues.
Single nucleotide polymorphism annotation is the process of predicting the effect or function of an individual SNP using SNP annotation tools. In SNP annotation the biological information is extracted, collected and displayed in a clear form amenable to query. SNP functional annotation is typically performed based on the available information on nucleic acid and protein sequences.

Coiled-coil domain containing 74A is a protein that in humans is encoded by the CCDC74A gene. The protein is most highly expressed in the testis and may play a role in developmental pathways. The gene has undergone duplication in the primate lineage within the last 9 million years, and its only true ortholog is found in Pan troglodytes.
References
- ↑ Smith LM, Kelleher NL (March 2018). "Proteoforms as the next proteomics currency". Science. 359 (6380): 1106–1107. Bibcode:2018Sci...359.1106S. doi:10.1126/science.aat1884. PMC 5944612 . PMID 29590032.
- ↑ Smith LM, Kelleher NL (March 2013). "Proteoform: a single term describing protein complexity". Nature Methods. 10 (3): 186–7. doi:10.1038/nmeth.2369. PMC 4114032 . PMID 23443629.
- ↑ Lima DB, Dupré M, Duchateau M, Gianetto QG, Rey M, Matondo M, Chamot-Rooke J (November 2020). Valencia A (ed.). "ProteoCombiner: integrating bottom-up with top-down proteomics data for improved proteoform assessment" (PDF). Bioinformatics: btaa958. doi:10.1093/bioinformatics/btaa958. PMID 33165572.
- ↑ Aebersold R, Agar JN, Amster IJ, Baker MS, Bertozzi CR, Boja ES, et al. (February 2018). "How many human proteoforms are there?". Nature Chemical Biology. 14 (3): 206–214. doi:10.1038/nchembio.2576. PMC 5837046 . PMID 29443976.