Related Research Articles
Molecular biology is a branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions.
In molecular biology, restriction fragment length polymorphism (RFLP) is a technique that exploits variations in homologous DNA sequences, known as polymorphisms, populations, or species or to pinpoint the locations of genes within a sequence. The term may refer to a polymorphism itself, as detected through the differing locations of restriction enzyme sites, or to a related laboratory technique by which such differences can be illustrated. In RFLP analysis, a DNA sample is digested into fragments by one or more restriction enzymes, and the resulting restriction fragments are then separated by gel electrophoresis according to their size.

Southern blot is a method used for detection and quantification of a specific DNA sequence in DNA samples. This method is used in molecular biology. Briefly, purified DNA from a biological sample is digested with restriction enzymes, and the resulting DNA fragments are separated by using an electric current to move them through a sieve-like gel or matrix, which allows smaller fragments to move faster than larger fragments. The DNA fragments are transferred out of the gel or matrix onto a solid membrane, which is then exposed to a DNA probe labeled with a radioactive, fluorescent, or chemical tag. The tag allows any DNA fragments containing complementary sequences with the DNA probe sequence to be visualized within the Southern blot.

Molecular genetics is a branch of biology that addresses how differences in the structures or expression of DNA molecules manifests as variation among organisms. Molecular genetics often applies an "investigative approach" to determine the structure and/or function of genes in an organism's genome using genetic screens.

DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter, DNA methylation typically acts to repress gene transcription. In mammals, DNA methylation is essential for normal development and is associated with a number of key processes including genomic imprinting, X-chromosome inactivation, repression of transposable elements, aging, and carcinogenesis.

An epigenome consists of a record of the chemical changes to the DNA and histone proteins of an organism; these changes can be passed down to an organism's offspring via transgenerational stranded epigenetic inheritance. Changes to the epigenome can result in changes to the structure of chromatin and changes to the function of the genome.
Genetics, a discipline of biology, is the science of heredity and variation in living organisms.

Amplified fragment length polymorphism is a PCR-based tool used in genetics research, DNA fingerprinting, and in the practice of genetic engineering. Developed in the early 1990s by KeyGene, AFLP uses restriction enzymes to digest genomic DNA, followed by ligation of adaptors to the sticky ends of the restriction fragments. A subset of the restriction fragments is then selected to be amplified. This selection is achieved by using primers complementary to the adaptor sequence, the restriction site sequence and a few nucleotides inside the restriction site fragments. The amplified fragments are separated and visualized on denaturing on agarose gel electrophoresis, either through autoradiography or fluorescence methodologies, or via automated capillary sequencing instruments.
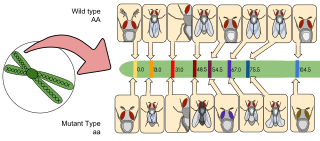
Gene mapping or genome mapping describes the methods used to identify the location of a gene on a chromosome and the distances between genes. Gene mapping can also describe the distances between different sites within a gene.
Ribotyping is a molecular technique for bacterial identification and characterization that uses information from rRNA-based phylogenetic analyses. It is a rapid and specific method widely used in clinical diagnostics and analysis of microbial communities in food, water, and beverages.
A genomic library is a collection of overlapping DNA fragments that together make up the total genomic DNA of a single organism. The DNA is stored in a population of identical vectors, each containing a different insert of DNA. In order to construct a genomic library, the organism's DNA is extracted from cells and then digested with a restriction enzyme to cut the DNA into fragments of a specific size. The fragments are then inserted into the vector using DNA ligase. Next, the vector DNA can be taken up by a host organism - commonly a population of Escherichia coli or yeast - with each cell containing only one vector molecule. Using a host cell to carry the vector allows for easy amplification and retrieval of specific clones from the library for analysis.

Bisulfitesequencing (also known as bisulphite sequencing) is the use of bisulfite treatment of DNA before routine sequencing to determine the pattern of methylation. DNA methylation was the first discovered epigenetic mark, and remains the most studied. In animals it predominantly involves the addition of a methyl group to the carbon-5 position of cytosine residues of the dinucleotide CpG, and is implicated in repression of transcriptional activity.
Population genomics is the large-scale comparison of DNA sequences of populations. Population genomics is a neologism that is associated with population genetics. Population genomics studies genome-wide effects to improve our understanding of microevolution so that we may learn the phylogenetic history and demography of a population.
Epigenomics is the study of the complete set of epigenetic modifications on the genetic material of a cell, known as the epigenome. The field is analogous to genomics and proteomics, which are the study of the genome and proteome of a cell. Epigenetic modifications are reversible modifications on a cell's DNA or histones that affect gene expression without altering the DNA sequence. Epigenomic maintenance is a continuous process and plays an important role in stability of eukaryotic genomes by taking part in crucial biological mechanisms like DNA repair. Plant flavones are said to be inhibiting epigenomic marks that cause cancers. Two of the most characterized epigenetic modifications are DNA methylation and histone modification. Epigenetic modifications play an important role in gene expression and regulation, and are involved in numerous cellular processes such as in differentiation/development and tumorigenesis. The study of epigenetics on a global level has been made possible only recently through the adaptation of genomic high-throughput assays.
The Illumina Methylation Assay using the Infinium I platform uses 'BeadChip' technology to generate a comprehensive genome-wide profiling of human DNA methylation. Similar to bisulfite sequencing and pyrosequencing, this method quantifies methylation levels at various loci within the genome. This assay is used for methylation probes on the Illumina Infinium HumanMethylation27 BeadChip. Probes on the 27k array target regions of the human genome to measure methylation levels at 27,578 CpG dinucleotides in 14,495 genes. In 2008, Illumina released the Infinium HumanMethylation450 BeadChip array, which targets over 450,000 methylation sites. In 2016, the Infinium MethylationEPIC BeadChip ("EPIC") was released, which interrogates over 850,000 methylation sites across the human genome.
Methylated DNA immunoprecipitation is a large-scale purification technique in molecular biology that is used to enrich for methylated DNA sequences. It consists of isolating methylated DNA fragments via an antibody raised against 5-methylcytosine (5mC). This technique was first described by Weber M. et al. in 2005 and has helped pave the way for viable methylome-level assessment efforts, as the purified fraction of methylated DNA can be input to high-throughput DNA detection methods such as high-resolution DNA microarrays (MeDIP-chip) or next-generation sequencing (MeDIP-seq). Nonetheless, understanding of the methylome remains rudimentary; its study is complicated by the fact that, like other epigenetic properties, patterns vary from cell-type to cell-type.

Combined Bisulfite Restriction Analysis is a molecular biology technique that allows for the sensitive quantification of DNA methylation levels at a specific genomic locus on a DNA sequence in a small sample of genomic DNA. The technique is a variation of bisulfite sequencing, and combines bisulfite conversion based polymerase chain reaction with restriction digestion. Originally developed to reliably handle minute amounts of genomic DNA from microdissected paraffin-embedded tissue samples, the technique has since seen widespread usage in cancer research and epigenetics studies.
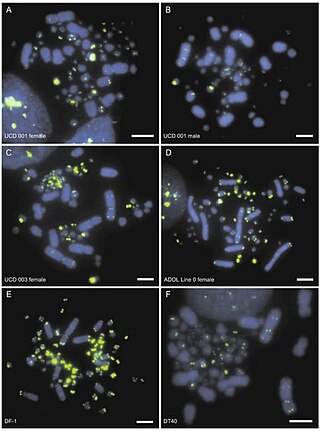
A mega-telomere, is an extremely long telomere sequence that sits on the end of chromosomes and prevents the loss of genetic information during cell replication. Like regular telomeres, mega-telomeres are made of a repetitive sequence of DNA and associated proteins, and are located on the ends of chromosomes. However, mega-telomeres are substantially longer than regular telomeres, ranging in size from 50 kilobases to several megabases.
Physical map is a technique used in molecular biology to find the order and physical distance between DNA base pairs by DNA markers. It is one of the gene mapping techniques which can determine the sequence of DNA base pairs with high accuracy. Genetic mapping, another approach of gene mapping, can provide markers needed for the physical mapping. However, as the former deduces the relative gene position by recombination frequencies, it is less accurate than the latter.
Long-range restriction mapping is an alternative genomic mapping technique to short-range, also called fine-scale mapping. Both forms utilize restriction enzymes in order to decipher the previously unknown order of DNA segments; the main difference between the two being the amount of DNA that comprises the final map. The unknown DNA is broken into many smaller fragments by these restriction enzymes at specific sites on the molecule, and then the fragments can later be analyzed by their individual sizes. A final long-range map can span hundreds to thousands of kilobytes of genetic data at many different loci.
References
- ↑ Costello, JF; Joseph F. Costello; Christoph Plass; Webster K. Cavenee (2002-03-01). "Restriction Landmark Genome Scanning". DNA Methylation Protocols. Methods in Molecular Biology. Vol. 200. pp. 53–70. doi:10.1385/1-59259-182-5:053. ISBN 1-59259-182-5. PMID 11951655.