Related Research Articles

Gene therapy is a medical technology that aims to produce a therapeutic effect through the manipulation of gene expression or through altering the biological properties of living cells.

A tumor suppressor gene (TSG), or anti-oncogene, is a gene that regulates a cell during cell division and replication. If the cell grows uncontrollably, it will result in cancer. When a tumor suppressor gene is mutated, it results in a loss or reduction in its function. In combination with other genetic mutations, this could allow the cell to grow abnormally. The loss of function for these genes may be even more significant in the development of human cancers, compared to the activation of oncogenes.

Thalassemias are inherited blood disorders that result in abnormal hemoglobin. Symptoms depend on the type of thalassemia and can vary from none to severe. Often there is mild to severe anemia as thalassemia can affect the production of red blood cells and also affect how long the red blood cells live. Symptoms of anemia include feeling tired and having pale skin. Other symptoms of thalassemia include bone problems, an enlarged spleen, yellowish skin, pulmonary hypertension, and dark urine. Slow growth may occur in children. Symptoms and presentations of thalassemia can change over time.

Molecular genetics is a branch of biology that addresses how differences in the structures or expression of DNA molecules manifests as variation among organisms. Molecular genetics often applies an "investigative approach" to determine the structure and/or function of genes in an organism's genome using genetic screens.
A genetic screen or mutagenesis screen is an experimental technique used to identify and select individuals who possess a phenotype of interest in a mutagenized population. Hence a genetic screen is a type of phenotypic screen. Genetic screens can provide important information on gene function as well as the molecular events that underlie a biological process or pathway. While genome projects have identified an extensive inventory of genes in many different organisms, genetic screens can provide valuable insight as to how those genes function.
In genetics, a nonsense mutation is a point mutation in a sequence of DNA that results in a nonsense codon, or a premature stop codon in the transcribed mRNA, and leads to a truncated, incomplete, and possibly nonfunctional protein product. Nonsense mutation is not always harmful, the functional effect of a nonsense mutation depends on many aspects, such as the location of the stop codon within the coding DNA. For example, the effect of a nonsense mutation depends on the proximity of the nonsense mutation to the original stop codon, and the degree to which functional subdomains of the protein are affected. As nonsense mutations leads to premature termination of polypeptide chains; they are also called chain termination mutations.

A germline mutation, or germinal mutation, is any detectable variation within germ cells. Mutations in these cells are the only mutations that can be passed on to offspring, when either a mutated sperm or oocyte come together to form a zygote. After this fertilization event occurs, germ cells divide rapidly to produce all of the cells in the body, causing this mutation to be present in every somatic and germline cell in the offspring; this is also known as a constitutional mutation. Germline mutation is distinct from somatic mutation.

Cystic fibrosis transmembrane conductance regulator (CFTR) is a membrane protein and anion channel in vertebrates that is encoded by the CFTR gene.

Copy number variation (CNV) is a phenomenon in which sections of the genome are repeated and the number of repeats in the genome varies between individuals. Copy number variation is a type of structural variation: specifically, it is a type of duplication or deletion event that affects a considerable number of base pairs. Approximately two-thirds of the entire human genome may be composed of repeats and 4.8–9.5% of the human genome can be classified as copy number variations. In mammals, copy number variations play an important role in generating necessary variation in the population as well as disease phenotype.
An insulator is a type of cis-regulatory element known as a long-range regulatory element. Found in multicellular eukaryotes and working over distances from the promoter element of the target gene, an insulator is typically 300 bp to 2000 bp in length. Insulators contain clustered binding sites for sequence specific DNA-binding proteins and mediate intra- and inter-chromosomal interactions.

Alpha-thalassemia is a form of thalassemia involving the genes HBA1 and HBA2. Thalassemias are a group of inherited blood conditions which result in the impaired production of hemoglobin, the molecule that carries oxygen in the blood. Normal hemoglobin consists of two alpha chains and two beta chains; in alpha-thalassemia, there is a quantitative decrease in the amount of alpha chains, resulting in fewer normal hemoglobin molecules. Furthermore, alpha-thalassemia leads to the production of unstable beta globin molecules which cause increased red blood cell destruction. The degree of impairment is based on which clinical phenotype is present.
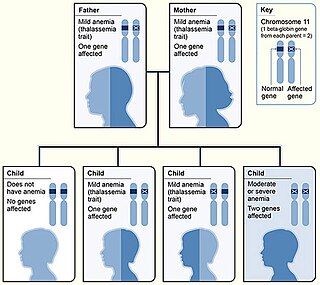
Beta thalassemias are a group of inherited blood disorders. They are forms of thalassemia caused by reduced or absent synthesis of the beta chains of hemoglobin that result in variable outcomes ranging from severe anemia to clinically asymptomatic individuals. Global annual incidence is estimated at one in 100,000. Beta thalassemias occur due to malfunctions in the hemoglobin subunit beta or HBB. The severity of the disease depends on the nature of the mutation.

Chromosome conformation capture techniques are a set of molecular biology methods used to analyze the spatial organization of chromatin in a cell. These methods quantify the number of interactions between genomic loci that are nearby in 3-D space, but may be separated by many nucleotides in the linear genome. Such interactions may result from biological functions, such as promoter-enhancer interactions, or from random polymer looping, where undirected physical motion of chromatin causes loci to collide. Interaction frequencies may be analyzed directly, or they may be converted to distances and used to reconstruct 3-D structures.

Hemoglobin subunit zeta is a protein that in humans is encoded by the HBZ gene.

Hemoglobin, alpha pseudogene 1, also known as HBAP1, is a human gene.

The 1000 Genomes Project, taken place from January 2008 to 2015, was an international research effort to establish the most detailed catalogue of human genetic variation at the time. Scientists planned to sequence the genomes of at least one thousand anonymous healthy participants from a number of different ethnic groups within the following three years, using advancements in newly developed technologies. In 2010, the project finished its pilot phase, which was described in detail in a publication in the journal Nature. In 2012, the sequencing of 1092 genomes was announced in a Nature publication. In 2015, two papers in Nature reported results and the completion of the project and opportunities for future research.
Douglas Roland Higgs FRS is a Professor of Molecular Haematology at the Weatherall Institute of Molecular Medicine, at the University of Oxford. He is known for his work on the regulation of alpha-globin and the genetics of alpha-thalassemia. He is currently working in understanding the mechanisms by which any mammalian gene is switched on and off during differentiation and development.
The history of genetics can be represented on a timeline of events from the earliest work in the 1850s, to the DNA era starting in the 1940s, and the genomics era beginning in the 1970s.
Johanna Rommens is a Canadian geneticist who was on the research team which identified and cloned the CFTR gene, which when mutated, is responsible for causing cystic fibrosis (CF). She later discovered the gene responsible for Shwachman-Diamond syndrome, a rare genetic disorder that causes pancreatic and hematologic problems. She is a Senior Scientist Emeritus at SickKids Research Institute and a professor in the Department of Molecular Genetics at the University of Toronto.
Haig H. Kazazian, Jr. was a professor in the Department of Genetic Medicine at Johns Hopkins University School of Medicine in Baltimore, Maryland. Kazazian was an elected member of the National Academy of Sciences and the American Academy of Arts and Sciences.
References
- ↑ Who's Who 2019. A & C Black, London. 2018. ISBN 978-1-472-94758-1.
- ↑ "Professor Bob Williamson". Murdoch Children's Research Institute. Retrieved 15 August 2018.
- ↑ Williamson, R. Properties of rapidly labelled deoxyribonucleic acid fragments isolated from the cytoplasm of primary cultures of embryonic mouse liver cells. J. Mol. Biol. 51, 157-160 (1970). https://doi.org/10.1016/0022-2836(70)90277-9
- ↑ Henikoff, S. & Church, G.M. Simultaneous Discovery of Cell-Free DNA and the Nucleosome Ladder. Genetics 209, 27-29 (2018). https://doi.org/10.1534/genetics.118.300775
- ↑ Ottolenghi, S. et al. Gene deletion as the cause of α thalassaemia: The severe form of α thalassaemia is caused by a haemoglobin gene deletion. Nature 251, 389-392 (1974). https://doi.org/10.1038/251389a0
- ↑ Ottolenghi, S. et al. δβ-Thalassemia is due to a gene deletion. Cell 9, 71-80 (1976). https://doi.org/10.1038/251389a0
- ↑ Murray, J.M. et al. Linkage relationship of a cloned DNA sequence on the short arm of the X chromosome to Duchenne muscular dystrophy. Nature 300, 69-71 (1982). https://doi.org/10.1038/300069a0
- ↑ Wainwright, B.J. et al. Localization of cystic fibrosis locus to human chromosome 7cen–q22. Nature 318, 384-385 (1985). https://doi.org/10.1038/318384a0
- ↑ Estivill, X. et al. A candidate for the cystic fibrosis locus isolated by selection for methylation-free islands. Nature 326, 840-846 (1987). https://doi.org/10.1038/326840a0
- ↑ Kevin Davies. The search for the cystic fibrosis gene. New Scientist 20 October 1989. https://www.newscientist.com/article/mg12416873-900/
- ↑ Lench, N. et al. Simple non-invasive method to obtain DNA for gene analysis. Lancet 331, 1356-1368 (1988). https://doi.org/10.1016/S0140-6736(88)92178-2
- ↑ Goate, A. et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature 349, 704-706 (1991). https://doi.org/10.1038/349704a0
- ↑ Williamson, B. Nature 298, 416-418 (1982). https://doi.org/10.1038/298416a0
- ↑ Caplen, N.J. et al. Nature Medicine 1, 39-46 (1995).
- ↑ Davies, K.E. The Long Journey from Diagnosis to Therapy. Ann. Rev. Genomics & Human Genet. 21, 1-13 (2020). https://doi.org/10.1146/annurev-genom-112019-083518
- ↑ Hardy, J. The discovery of Alzheimer-causing mutations in the APP gene and the formulation of the “amyloid cascade hypothesis”. FEBS Journal 284, 1040-1044 (2017). https://doi.org/10.1111/febs.14004
- ↑ Choo, K.H.A. "David M. Danks, M.D., A.O. (June 4, 1931–July 8, 2003): Founder, Murdoch Childrens Research Institute". American Journal of Human Genetics 73: 981–985 (2003). doi:10.1086/379383.
- ↑ Wilton, L., Williamson, R., McBain J., Edgar, D., and Voullaire, L. “Birth of a Healthy Infant after Preimplantation Confirmation of Euploidy by Comparative Genomic Hybridisation” N. Engl. J. Med, 345:1537-1541 (2001).
- ↑ Delatycki, M.B., Williamson, R. and Forrest, S.M. “Friedreich Ataxia; an overview.” J. Med. Genet. 37:1-8 (2000).
- ↑ Collins, V., Halliday, J., Kahler, S., and Williamson, R. “Parents’ Experience with Genetic Counseling After the Birth of a Baby with a Genetic Disorder: An Exploratory Study.” J. Genet. Coun. 10:53-72 (2001).
- ↑ Dodson, M. and Williamson, R. “Indigenous People and the Morality of the Human Genome Diversity Project.” J. Med. Ethics 25:204-208 (1999).
- 1 2 "Robert Williamson". Royal Society. 12 August 2015. Retrieved 12 August 2018.
- ↑ Williamson, R. Universal community carrier screening for cystic fibrosis? Nature Genetics 3, 195-201 (1993). https://doi.org/10.1038/ng0393-195
- ↑ Williamson, R. & Duncan, R. DNA testing for all. Nature 418, 585-586 (2002). https://doi.org/10.1038/418585a
- ↑ "Professor Robert Williamson". King Faisal Prize. 10 October 2012. Retrieved 12 August 2018.