Alternative splicing, or alternative RNA splicing, or differential splicing, is a regulated process during gene expression that results in a single gene coding for multiple proteins. In this process, particular exons of a gene may be included within or excluded from the final, processed messenger RNA (mRNA) produced from that gene. Consequently, the proteins translated from alternatively spliced mRNAs will contain differences in their amino acid sequence and, often, in their biological functions. Notably, alternative splicing allows the human genome to direct the synthesis of many more proteins than would be expected from its 20,000 protein-coding genes.

A spliceosome is a large and complex molecular RNA identity found primarily within the nucleus of eukaryotic cells. The spliceosome is assembled from small nuclear RNAs (snRNA) and approximately 80 proteins. The spliceosome removes introns from a transcribed pre-mRNA, a type of primary transcript. This process is generally referred to as splicing. An analogy is a film editor, who selectively cuts out irrelevant or incorrect material from the initial film and sends the cleaned-up version to the director for the final cut.

SR proteins are a conserved family of proteins involved in RNA splicing. SR proteins are named because they contain a protein domain with long repeats of serine and arginine amino acid residues, whose standard abbreviations are "S" and "R" respectively. SR proteins are ~200-600 amino acids in length and composed of two domains, the RNA recognition motif (RRM) region and the RS domain. SR proteins are more commonly found in the nucleus than the cytoplasm, but several SR proteins are known to shuttle between the nucleus and the cytoplasm.
RNA-binding proteins are proteins that bind to the double or single stranded RNA in cells and participate in forming ribonucleoprotein complexes. RBPs contain various structural motifs, such as RNA recognition motif (RRM), dsRNA binding domain, zinc finger and others. They are cytoplasmic and nuclear proteins. However, since most mature RNA is exported from the nucleus relatively quickly, most RBPs in the nucleus exist as complexes of protein and pre-mRNA called heterogeneous ribonucleoprotein particles (hnRNPs). RBPs have crucial roles in various cellular processes such as: cellular function, transport and localization. They especially play a major role in post-transcriptional control of RNAs, such as: splicing, polyadenylation, mRNA stabilization, mRNA localization and translation. Eukaryotic cells encode diverse RBPs, approximately 500 genes, with unique RNA-binding activity and protein–protein interaction. During evolution, the diversity of RBPs greatly increased with the increase in the number of introns. Diversity enabled eukaryotic cells to utilize RNA exons in various arrangements, giving rise to a unique RNP (ribonucleoprotein) for each RNA. Although RBPs have a crucial role in post-transcriptional regulation in gene expression, relatively few RBPs have been studied systematically.
Small nuclear RNA (snRNA) is a class of small RNA molecules that are found within the splicing speckles and Cajal bodies of the cell nucleus in eukaryotic cells. The length of an average snRNA is approximately 150 nucleotides. They are transcribed by either RNA polymerase II or RNA polymerase III. Their primary function is in the processing of pre-messenger RNA (hnRNA) in the nucleus. They have also been shown to aid in the regulation of transcription factors or RNA polymerase II, and maintaining the telomeres.
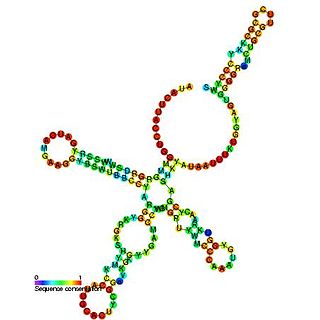
U1 spliceosomal RNA is the small nuclear RNA (snRNA) component of U1 snRNP, an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification, and takes place only in the nucleus of eukaryotes.

snRNP70 also known as U1 small nuclear ribonucleoprotein 70 kDa is a protein that in humans is encoded by the SNRNP70 gene. snRNP70 is a small nuclear ribonucleoprotein that associates with U1 spliceosomal RNA, forming the U1snRNP a core component of the spliceosome. The U1-70K protein and other components of the spliceosome complex form detergent-insoluble aggregates in both sporadic and familial human cases of Alzheimer's disease. U1-70K co-localizes with Tau in neurofibrillary tangles in Alzheimer's disease.

Splicing factor, proline- and glutamine-rich is a protein that in humans is encoded by the SFPQ gene.

PRP31 pre-mRNA processing factor 31 homolog , also known as PRPF31, is a protein which in humans is encoded by the PRPF31 gene.

RNA-binding protein with serine-rich domain 1 is a protein that in humans is encoded by the RNPS1 gene.

U4/U6.U5 tri-snRNP-associated protein 1 is a protein that in humans is encoded by the SART1 gene. This gene encodes two proteins, the SART1(800) protein expressed in the nucleus of the majority of proliferating cells, and the SART1(259) protein expressed in the cytosol of epithelial cancers. The SART1(259) protein is translated by the mechanism of -1 frameshifting during posttranscriptional regulation. The two encoded proteins are thought to be involved in the regulation of proliferation. Both proteins have tumor-rejection antigens. The SART1(259) protein possesses tumor epitopes capable of inducing HLA-A2402-restricted cytotoxic T lymphocytes in cancer patients. This SART1(259) antigen may be useful in specific immunotherapy for cancer patients and may serve as a paradigmatic tool for the diagnosis and treatment of patients with atopy. The SART1(259) protein is found to be essential for the recruitment of the tri-snRNP to the pre-spliceosome in the spliceosome assembly pathway.

Splicing factor 3A subunit 3 is a protein that in humans is encoded by the SF3A3 gene.

Heterogeneous nuclear ribonucleoprotein L is a protein that in humans is encoded by the HNRNPL gene.

Splicing factor 3B subunit 4 is a protein that in humans is encoded by the SF3B4 gene.

U2 small nuclear ribonucleoprotein B is a protein that in humans is encoded by the SNRPB2 gene.

U4/U6 small nuclear ribonucleoprotein Prp4 is a protein that in humans is encoded by the PRPF4 gene. The removal of introns from nuclear pre-mRNAs occurs on complexes called spliceosomes, which are made up of 4 small nuclear ribonucleoprotein (snRNP) particles and an undefined number of transiently associated splicing factors. PRPF4 is 1 of several proteins that associate with U4 and U6 snRNPs.[supplied by OMIM]
U1 small nuclear ribonucleoprotein C is a protein that in humans is encoded by the SNRPC gene.

Dermatan-sulfate epimerase is an enzyme that in humans is encoded by the DSE gene.

Serine/arginine-rich splicing factor 1 (SRSF1) also known as alternative splicing factor 1 (ASF1), pre-mRNA-splicing factor SF2 (SF2) or ASF1/SF2 is a protein that in humans is encoded by the SRSF1 gene. ASF/SF2 is an essential sequence specific splicing factor involved in pre-mRNA splicing. SRSF1 is the gene that codes for ASF/SF2 and is found on chromosome 17. The resulting splicing factor is a protein of approximately 33 kDa. ASF/SF2 is necessary for all splicing reactions to occur, and influences splice site selection in a concentration-dependent manner, resulting in alternative splicing. In addition to being involved in the splicing process, ASF/SF2 also mediates post-splicing activities, such as mRNA nuclear export and translation.
Prp24 is a protein part of the pre-messenger RNA splicing process and aids the binding of U6 snRNA to U4 snRNA during the formation of spliceosomes. Found in eukaryotes from yeast to E. coli, fungi, and humans, Prp24 was initially discovered to be an important element of RNA splicing in 1989. Mutations in Prp24 were later discovered in 1991 to suppress mutations in U4 that resulted in cold-sensitive strains of yeast, indicating its involvement in the reformation of the U4/U6 duplex after the catalytic steps of splicing.