
The endoplasmic reticulum (ER) is, in essence, the transportation system of the eukaryotic cell, and has many other important functions such as protein folding. It is a type of organelle made up of two subunits – rough endoplasmic reticulum (RER), and smooth endoplasmic reticulum (SER). The endoplasmic reticulum is found in most eukaryotic cells and forms an interconnected network of flattened, membrane-enclosed sacs known as cisternae, and tubular structures in the SER. The membranes of the ER are continuous with the outer nuclear membrane. The endoplasmic reticulum is not found in red blood cells, or spermatozoa.

The endomembrane system is composed of the different membranes (endomembranes) that are suspended in the cytoplasm within a eukaryotic cell. These membranes divide the cell into functional and structural compartments, or organelles. In eukaryotes the organelles of the endomembrane system include: the nuclear membrane, the endoplasmic reticulum, the Golgi apparatus, lysosomes, vesicles, endosomes, and plasma (cell) membrane among others. The system is defined more accurately as the set of membranes that forms a single functional and developmental unit, either being connected directly, or exchanging material through vesicle transport. Importantly, the endomembrane system does not include the membranes of plastids or mitochondria, but might have evolved partially from the actions of the latter.
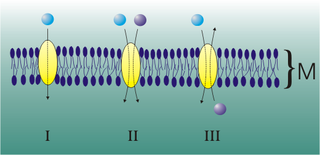
Mediated transport refers to transport mediated by a membrane transport protein. Substances in the human body may be hydrophobic, electrophilic, contain a positively or negatively charge, or have another property. As such there are times when those substances may not be able to pass over the cell membrane using protein-independent movement. The cell membrane is imbedded with many membrane transport proteins that allow such molecules to travel in and out of the cell. There are three types of mediated transporters: uniport, symport, and antiport. Things that can be transported are nutrients, ions, glucose, etc, all depending on the needs of the cell. One example of a uniport mediated transport protein is GLUT1. GLUT1 is a transmembrane protein, which means it spans the entire width of the cell membrane, connecting the extracellular and intracellular region. It is a uniport system because it specifically transports glucose in only one direction, down its concentration gradient across the cell membrane.
Calcium release-activated channels (CRAC) are specialized plasma membrane Ca2+ ion channels. When calcium ions (Ca2+) are depleted from the endoplasmic reticulum (a major store of Ca2+) of mammalian cells, the CRAC channel is activated to slowly replenish the level of calcium in the endoplasmic reticulum. The Ca2+ Release-activated Ca2+ (CRAC) Channel (CRAC-C) Family (TC# 1.A.52) is a member of the Cation Diffusion Facilitator (CDF) Superfamily. These proteins typically have between 4 and 6 transmembrane α-helical spanners (TMSs). The 4 TMS CRAC channels arose by loss of 2TMSs from 6TMS CDF carriers, an example of 'reverse' evolution'.
Voltage-gated calcium channels (VGCCs), also known as voltage-dependent calcium channels (VDCCs), are a group of voltage-gated ion channels found in the membrane of excitable cells (e.g., muscle, glial cells, neurons, etc.) with a permeability to the calcium ion Ca2+. These channels are slightly permeable to sodium ions, so they are also called Ca2+–Na+ channels, but their permeability to calcium is about 1000-fold greater than to sodium under normal physiological conditions.
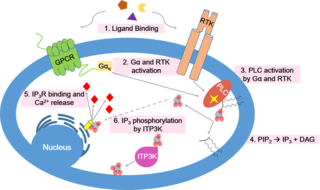
Calcium signaling is the use of calcium ions (Ca2+) to communicate and drive intracellular processes often as a step in signal transduction. Ca2+ is important for cellular signalling, for once it enters the cytosol of the cytoplasm it exerts allosteric regulatory effects on many enzymes and proteins. Ca2+ can act in signal transduction resulting from activation of ion channels or as a second messenger caused by indirect signal transduction pathways such as G protein-coupled receptors.
The sodium-calcium exchanger (often denoted Na+/Ca2+ exchanger, exchange protein, or NCX) is an antiporter membrane protein that removes calcium from cells. It uses the energy that is stored in the electrochemical gradient of sodium (Na+) by allowing Na+ to flow down its gradient across the plasma membrane in exchange for the countertransport of calcium ions (Ca2+). A single calcium ion is exported for the import of three sodium ions. The exchanger exists in many different cell types and animal species. The NCX is considered one of the most important cellular mechanisms for removing Ca2+.

Ca2+ ATPase is a form of P-ATPase that transfers calcium after a muscle has contracted. The two kinds of calcium ATPase are:

TAP-associated glycoprotein, also known as tapasin or TAPBP, is a protein that in humans is encoded by the TAPBP gene.

Calcium release-activated calcium channel protein 1 is a calcium selective ion channel that in humans is encoded by the ORAI1 gene. Orai channels play an important role in the activation of T-lymphocytes. The loss of function mutation of Orai1 causes severe combined immunodeficiency (SCID) in humans The mammalian orai family has two additional homologs, Orai2 and Orai3. Orai proteins share no homology with any other ion channel family of any other known proteins. They have 4 transmembrane domains and form hexamers.
Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (SERCA1) also known as Calcium pump 1, is an enzyme that in humans is encoded by the ATP2A1 gene.

Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 is an enzyme that in humans is encoded by the ATP2A3 gene.

Stromal interaction molecule 2 (STIM2) is a protein that in humans is encoded by the STIM2 gene.

SARAF is a protein that in humans is encoded by the SARAF gene, formerly known as TMEM66.
Membrane contact sites (MCS) are close appositions between two organelles. Ultrastructural studies typically reveal an intermembrane distance in the order of the size of a single protein, as small as 10 nm or wider, with no clear upper limit. These zones of apposition are highly conserved in evolution. These sites are thought to be important to facilitate signalling, and they promote the passage of small molecules, including ions, lipids and reactive oxygen species. MCS are important in the function of the endoplasmic reticulum (ER), since this is the major site of lipid synthesis within cells. The ER makes close contact with many organelles, including mitochondria, Golgi, endosomes, lysosomes, peroxisomes, chloroplasts and the plasma membrane. Both mitochondria and sorting endosomes undergo major rearrangements leading to fission where they contact the ER. Sites of close apposition can also form between most of these organelles most pairwise combinations. First mentions of these contact sites can be found in papers published in the late 1950s mainly visualized using electron microscopy (EM) techniques. Copeland and Dalton described them as “highly specialized tubular form of endoplasmic reticulum in association with the mitochondria and apparently in turn, with the vascular border of the cell”.
LiMETER stands for light-inducible membrane-tethered peripheral endoplasmic reticulum (ER). LiMETER is an optogenetics tool designed to reversibly label cortical ER or the apposition between plasma membrane (PM) and endoplasmic reticulum (ER) membranes.

GRAM domain containing 1C also known as Aster-C is a cholesterol transport protein that is encoded by the GRAMD1C gene. It contains a transmembrane region, a GRAM domain and a VASt domain. It is anchored to the endoplasmic reticulum through its transmembrane domain.

GRAM domain-containing 2A protein is a protein encoded by the GRAMD2A gene. Like GRAMD2B, the protein consists of a GRAM domain and a transmembrane domain that anchors it to the endoplasmic reticulum.
Patrick G. Hogan is a cellular and molecular biologist who studies how cellular signaling leads to gene expression. He obtained his bachelor’s degree from Harvard University and a PhD in neurobiology from Harvard Medical School. In 2010, he moved to the La Jolla Institute for Immunology in San Diego as a Professor in the Division of Signaling and Gene Expression. He is a Founder and Member of the Scientific Advisory Board, CalciMedica Inc, La Jolla, CA.

















