
Inbreeding is the production of offspring from the mating or breeding of individuals or organisms that are closely related genetically. By analogy, the term is used in human reproduction, but more commonly refers to the genetic disorders and other consequences that may arise from expression of deleterious or recessive traits resulting from incestuous sexual relationships and consanguinity. Animals avoid incest only rarely.

Fitness is the quantitative representation of individual reproductive success. It is also equal to the average contribution to the gene pool of the next generation, made by the same individuals of the specified genotype or phenotype. Fitness can be defined either with respect to a genotype or to a phenotype in a given environment or time. The fitness of a genotype is manifested through its phenotype, which is also affected by the developmental environment. The fitness of a given phenotype can also be different in different selective environments.
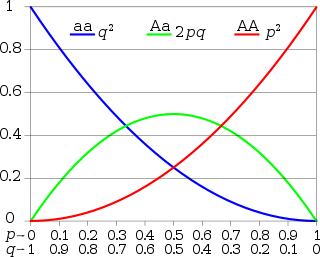
In population genetics, the Hardy–Weinberg principle, also known as the Hardy–Weinberg equilibrium, model, theorem, or law, states that allele and genotype frequencies in a population will remain constant from generation to generation in the absence of other evolutionary influences. These influences include genetic drift, mate choice, assortative mating, natural selection, sexual selection, mutation, gene flow, meiotic drive, genetic hitchhiking, population bottleneck, founder effect,inbreeding and outbreeding depression.

Quantitative genetics deals with phenotypes that vary continuously —as opposed to discretely identifiable phenotypes and gene-products.
In statistics, Gibbs sampling or a Gibbs sampler is a Markov chain Monte Carlo (MCMC) algorithm for obtaining a sequence of observations which are approximated from a specified multivariate probability distribution, when direct sampling is difficult. This sequence can be used to approximate the joint distribution ; to approximate the marginal distribution of one of the variables, or some subset of the variables ; or to compute an integral. Typically, some of the variables correspond to observations whose values are known, and hence do not need to be sampled.
The coefficient of relationship is a measure of the degree of consanguinity between two individuals. The term coefficient of relationship was defined by Sewall Wright in 1922, and was derived from his definition of the coefficient of inbreeding of 1921. The measure is most commonly used in genetics and genealogy. A coefficient of inbreeding can be calculated for an individual, and is typically one-half the coefficient of relationship between the parents.
In population genetics, F-statistics describe the statistically expected level of heterozygosity in a population; more specifically the expected degree of (usually) a reduction in heterozygosity when compared to Hardy–Weinberg expectation.
In electrical engineering, statistical computing and bioinformatics, the Baum–Welch algorithm is a special case of the expectation–maximization algorithm used to find the unknown parameters of a hidden Markov model (HMM). It makes use of the forward-backward algorithm to compute the statistics for the expectation step.

A seed orchard is an intensively-managed plantation of specifically arranged trees for the mass production of genetically improved seeds to create plants, or seeds for the establishment of new forests.
The effective population size (Ne) is a number that, in some simplified scenarios, corresponds to the number of breeding individuals in the population. More generally, Ne is the number of individuals that an idealised population would need to have in order for some specified quantity of interest to be the same as in the real population. Idealised populations are based on unrealistic but convenient simplifications such as random mating, simultaneous birth of each new generation, constant population size, and equal numbers of children per parent. For most quantities of interest and most real populations, the effective population size Ne is usually smaller than the census population size N of a real population. The same population may have multiple effective population sizes, for different properties of interest, including for different genetic loci.

A DNA segment is identical by state (IBS) in two or more individuals if they have identical nucleotide sequences in this segment. An IBS segment is identical by descent (IBD) in two or more individuals if they have inherited it from a common ancestor without recombination, that is, the segment has the same ancestral origin in these individuals. DNA segments that are IBD are IBS per definition, but segments that are not IBD can still be IBS due to the same mutations in different individuals or recombinations that do not alter the segment.
Genetic load is the difference between the fitness of an average genotype in a population and the fitness of some reference genotype, which may be either the best present in a population, or may be the theoretically optimal genotype. The average individual taken from a population with a low genetic load will generally, when grown in the same conditions, have more surviving offspring than the average individual from a population with a high genetic load. Genetic load can also be seen as reduced fitness at the population level compared to what the population would have if all individuals had the reference high-fitness genotype. High genetic load may put a population in danger of extinction.

Genetic distance is a measure of the genetic divergence between species or between populations within a species, whether the distance measures time from common ancestor or degree of differentiation. Populations with many similar alleles have small genetic distances. This indicates that they are closely related and have a recent common ancestor.
Malecot's coancestry coefficient, , refers to an indirect measure of genetic similarity of two individuals which was initially devised by the French mathematician Gustave Malécot.
In population genetics, fixation is the change in a gene pool from a situation where there exists at least two variants of a particular gene (allele) in a given population to a situation where only one of the alleles remains. In the absence of mutation or heterozygote advantage, any allele must eventually be lost completely from the population or fixed. Whether a gene will ultimately be lost or fixed is dependent on selection coefficients and chance fluctuations in allelic proportions. Fixation can refer to a gene in general or particular nucleotide position in the DNA chain (locus).
A doubled haploid (DH) is a genotype formed when haploid cells undergo chromosome doubling. Artificial production of doubled haploids is important in plant breeding.

Zygosity is the degree to which both copies of a chromosome or gene have the same genetic sequence. In other words, it is the degree of similarity of the alleles in an organism.

Isolation by distance (IBD) is a term used to refer to the accrual of local genetic variation under geographically limited dispersal. The IBD model is useful for determining the distribution of gene frequencies over a geographic region. Both dispersal variance and migration probabilities are variables in this model and both contribute to local genetic differentiation. Isolation by distance is usually the simplest model for the cause of genetic isolation between populations. Evolutionary biologists and population geneticists have been exploring varying theories and models for explaining population structure. Yoichi Ishida compares two important theories of isolation by distance and clarifies the relationship between the two. According to Ishida, Sewall Wright's isolation by distance theory is termed ecological isolation by distance while Gustave Malécot's theory is called genetic isolation by distance. Isolation by distance is distantly related to speciation. Multiple types of isolating barriers, namely prezygotic isolating barriers, including isolation by distance, are considered the key factor in keeping populations apart, limiting gene flow.
In probability theory and directional statistics, a wrapped probability distribution is a continuous probability distribution that describes data points that lie on a unit n-sphere. In one dimension, a wrapped distribution consists of points on the unit circle. If is a random variate in the interval with probability density function (PDF) , then is a circular variable distributed according to the wrapped distribution and is an angular variable in the interval distributed according to the wrapped distribution .
Genetic purging is the reduction of the frequency of a deleterious allele, caused by an increased efficiency of natural selection prompted by inbreeding.










