
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA. The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as regulatory sequences, and often a substantial fraction of junk DNA with no evident function. Almost all eukaryotes have mitochondria and a small mitochondrial genome. Algae and plants also contain chloroplasts with a chloroplast genome.

Genomics is an interdisciplinary field of biology focusing on the structure, function, evolution, mapping, and editing of genomes. A genome is an organism's complete set of DNA, including all of its genes as well as its hierarchical, three-dimensional structural configuration. In contrast to genetics, which refers to the study of individual genes and their roles in inheritance, genomics aims at the collective characterization and quantification of all of an organism's genes, their interrelations and influence on the organism. Genes may direct the production of proteins with the assistance of enzymes and messenger molecules. In turn, proteins make up body structures such as organs and tissues as well as control chemical reactions and carry signals between cells. Genomics also involves the sequencing and analysis of genomes through uses of high throughput DNA sequencing and bioinformatics to assemble and analyze the function and structure of entire genomes. Advances in genomics have triggered a revolution in discovery-based research and systems biology to facilitate understanding of even the most complex biological systems such as the brain.

Selaginella is the sole genus in the family Selaginellaceae, the spikemosses or lesser clubmosses, a kind of vascular plant.

Mitochondrial DNA is the DNA located in mitochondria, cellular organelles within eukaryotic cells that convert chemical energy from food into a form that cells can use, such as adenosine triphosphate (ATP). Mitochondrial DNA is only a small portion of the DNA in a eukaryotic cell; most of the DNA can be found in the cell nucleus and, in plants and algae, also in plastids such as chloroplasts.
Molecular evolution is the process of change in the sequence composition of cellular molecules such as DNA, RNA, and proteins across generations. The field of molecular evolution uses principles of evolutionary biology and population genetics to explain patterns in these changes. Major topics in molecular evolution concern the rates and impacts of single nucleotide changes, neutral evolution vs. natural selection, origins of new genes, the genetic nature of complex traits, the genetic basis of speciation, the evolution of development, and ways that evolutionary forces influence genomic and phenotypic changes.

Comparative genomics is a field of biological research in which the genomic features of different organisms are compared. The genomic features may include the DNA sequence, genes, gene order, regulatory sequences, and other genomic structural landmarks. In this branch of genomics, whole or large parts of genomes resulting from genome projects are compared to study basic biological similarities and differences as well as evolutionary relationships between organisms. The major principle of comparative genomics is that common features of two organisms will often be encoded within the DNA that is evolutionarily conserved between them. Therefore, comparative genomic approaches start with making some form of alignment of genome sequences and looking for orthologous sequences in the aligned genomes and checking to what extent those sequences are conserved. Based on these, genome and molecular evolution are inferred and this may in turn be put in the context of, for example, phenotypic evolution or population genetics.

Genome size is the total amount of DNA contained within one copy of a single complete genome. It is typically measured in terms of mass in picograms or less frequently in daltons, or as the total number of nucleotide base pairs, usually in megabases. One picogram is equal to 978 megabases. In diploid organisms, genome size is often used interchangeably with the term C-value.

A resurrection plant is any poikilohydric plant that can survive extreme dehydration, even over months or years.

The Joint Genome Institute (JGI) is a scientific user facility for integrative genomic science at Lawrence Berkeley National Laboratory. The mission of the JGI is to advance genomics research in support of the United States Department of Energy's (DOE) missions of energy and the environment. It is one of three national scientific user facilities supported by the Office of Biological and Environmental Research (BER) within the Department of Energy's Office of Research. These BER facilities are part of a more extensive network of 28 national scientific user facilities that operate at the DOE national laboratories.

The Human Genome Project (HGP) was an international scientific research project with the goal of determining the base pairs that make up human DNA, and of identifying, mapping and sequencing all of the genes of the human genome from both a physical and a functional standpoint. It started in 1990 and was completed in 2003. It remains the world's largest collaborative biological project. Planning for the project started after it was adopted in 1984 by the US government, and it officially launched in 1990. It was declared complete on April 14, 2003, and included about 92% of the genome. Level "complete genome" was achieved in May 2021, with a remaining only 0.3% bases covered by potential issues. The final gapless assembly was finished in January 2022.
The CoRR hypothesis states that the location of genetic information in cytoplasmic organelles permits regulation of its expression by the reduction-oxidation ("redox") state of its gene products.

In molecular biology and genetics, DNA annotation or genome annotation is the process of describing the structure and function of the components of a genome, by analyzing and interpreting them in order to extract their biological significance and understand the biological processes in which they participate. Among other things, it identifies the locations of genes and all the coding regions in a genome and determines what those genes do.

Gloria M. Coruzzi is an American molecular biologist specializing in plant systems biology and evolutionary genomics.
Deinococcus frigens is a species of low temperature and drought-tolerating, UV-resistant bacteria from Antarctica. It is Gram-positive, non-motile and coccoid-shaped. Its type strain is AA-692. Individual Deinococcus frigens range in size from 0.9-2.0 μm and colonies appear orange or pink in color. Liquid-grown cells viewed using phase-contrast light microscopy and transmission electron microscopy on agar-coated slides show that isolated D. frigens appear to produce buds. Comparison of the genomes of Deiococcus radiodurans and D. frigens have predicted that no flagellar assembly exists in D. frigens.
Nikos Kyrpides is a Greek-American bioscientist who has worked on the origins of life, information processing, bioinformatics, microbiology, metagenomics and microbiome data science. He is a senior staff scientist at the Berkeley National Laboratory, head of the Prokaryote Super Program and leads the Microbiome Data Science program at the US Department of Energy Joint Genome Institute.
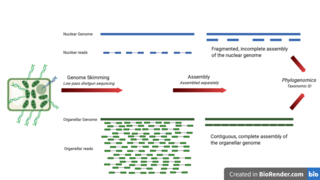
Genome skimming is a sequencing approach that uses low-pass, shallow sequencing of a genome, to generate fragments of DNA, known as genome skims. These genome skims contain information about the high-copy fraction of the genome. The high-copy fraction of the genome consists of the ribosomal DNA, plastid genome (plastome), mitochondrial genome (mitogenome), and nuclear repeats such as microsatellites and transposable elements. It employs high-throughput, next generation sequencing technology to generate these skims. Although these skims are merely 'the tip of the genomic iceberg', phylogenomic analysis of them can still provide insights on evolutionary history and biodiversity at a lower cost and larger scale than traditional methods. Due to the small amount of DNA required for genome skimming, its methodology can be applied in other fields other than genomics. Tasks like this include determining the traceability of products in the food industry, enforcing international regulations regarding biodiversity and biological resources, and forensics.












