Related Research Articles

In genetics and bioinformatics, a single-nucleotide polymorphism is a germline substitution of a single nucleotide at a specific position in the genome that is present in a sufficiently large fraction of considered population.
The International HapMap Project was an organization that aimed to develop a haplotype map (HapMap) of the human genome, to describe the common patterns of human genetic variation. HapMap is used to find genetic variants affecting health, disease and responses to drugs and environmental factors. The information produced by the project is made freely available for research.

A DNA segment is identical by state (IBS) in two or more individuals if they have identical nucleotide sequences in this segment. An IBS segment is identical by descent (IBD) in two or more individuals if they have inherited it from a common ancestor without recombination, that is, the segment has the same ancestral origin in these individuals. DNA segments that are IBD are IBS per definition, but segments that are not IBD can still be IBS due to the same mutations in different individuals or recombinations that do not alter the segment.

Human genetic variation is the genetic differences in and among populations. There may be multiple variants of any given gene in the human population (alleles), a situation called polymorphism.
David Emil Reich is an American geneticist known for his research into the population genetics of ancient humans, including their migrations and the mixing of populations, discovered by analysis of genome-wide patterns of mutations. He is professor in the department of genetics at the Harvard Medical School, and an associate of the Broad Institute. Reich was highlighted as one of Nature's 10 for his contributions to science in 2015. He received the Dan David Prize in 2017, the NAS Award in Molecular Biology, the Wiley Prize, and the Darwin–Wallace Medal in 2019. In 2021 he was awarded the Massry Prize.
Population structure is the presence of a systematic difference in allele frequencies between subpopulations. In a randomly mating population, allele frequencies are expected to be roughly similar between groups. However, mating tends to be non-random to some degree, causing structure to arise. For example, a barrier like a river can separate two groups of the same species and make it difficult for potential mates to cross; if a mutation occurs, over many generations it can spread and become common in one subpopulation while being completely absent in the other.

The Denisovans or Denisova hominins(di-NEE-sə-və) are an extinct species or subspecies of archaic human that ranged across Asia during the Lower and Middle Paleolithic. Denisovans are known from few physical remains; consequently, most of what is known about them comes from DNA evidence. No formal species name has been established pending more complete fossil material.
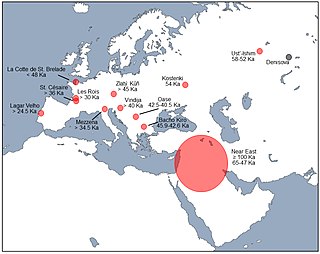
There is evidence for interbreeding between archaic and modern humans during the Middle Paleolithic and early Upper Paleolithic. The interbreeding happened in several independent events that included Neanderthals and Denisovans, as well as several unidentified hominins.
In genetics, haplotype estimation refers to the process of statistical estimation of haplotypes from genotype data. The most common situation arises when genotypes are collected at a set of polymorphic sites from a group of individuals. For example in human genetics, genome-wide association studies collect genotypes in thousands of individuals at between 200,000-5,000,000 SNPs using microarrays. Haplotype estimation methods are used in the analysis of these datasets and allow genotype imputation of alleles from reference databases such as the HapMap Project and the 1000 Genomes Project.
Imputation in genetics refers to the statistical inference of unobserved genotypes. It is achieved by using known haplotypes in a population, for instance from the HapMap or the 1000 Genomes Project in humans, thereby allowing to test for association between a trait of interest and experimentally untyped genetic variants, but whose genotypes have been statistically inferred ("imputed"). Genotype imputation is usually performed on SNPs, the most common kind of genetic variation.
Mega2 allows the applied statistical geneticist to convert one's data from several input formats to a large number output formats suitable for analysis by commonly used software packages. In a typical human genetics study, the analyst often needs to use a variety of different software programs to analyze the data, and these programs usually require that the data be formatted to their precise input specifications. Conversion of one's data into these multiple different formats can be tedious, time-consuming, and error-prone. Mega2, by providing validated conversion pipelines, can accelerate the analyses while reducing errors.

Johannes Krause is a German biochemist with a research focus on historical infectious diseases and human evolution. Since 2010, he has been professor of archaeology and paleogenetics at the University of Tübingen. In 2014, Krause was named a founding co-director of the new Max Planck Institute for the Science of Human History in Jena.
A ghost population is a population that has been inferred through using statistical techniques.

Jonathan Laurence Marchini is a Bayesian statistician and professor of statistical genomics in the Department of Statistics at the University of Oxford, a tutorial fellow in statistics at Somerville College, Oxford and a co-founder and director of Gensci Ltd. He co-leads the Haplotype Reference Consortium.

Denny is a ~90,000 year old fossil specimen belonging to a ~13-year-old Neanderthal-Denisovan hybrid girl. To date, she is the only first-generation hybrid hominin ever discovered. Denny’s remains consist of a single fossilized fragment of a long bone discovered among over 2,000 visually unidentifiable fragments excavated at the Denisova Cave in the Altai Mountains, Russia in 2012.
Sarah Anne Tishkoff is an American geneticist and the David and Lyn Silfen Professor in the Department of Genetics and Biology at the University of Pennsylvania. She also serves as a director for the American Society of Human Genetics and is an associate editor at PLOS Genetics, G3, and Genome Research. She is also a member of the scientific advisory board at the David and Lucile Packard Foundation.
Viviane Slon is a paleogeneticist at the Max Planck Institute for Evolutionary Anthropology. She identified that a teenage girl born 90,000 years ago had both Neanderthal and Denisovan parents. She was selected as one of Nature's 10 in 2018.
Amanda M. Hulse-Kemp is a computational biologist with the United States Department of Agriculture – Agricultural Research Service. She works in the Genomics and Bioinformatics Research Unit and is stationed on the North Carolina State University campus in Raleigh, North Carolina.
Human genetic clustering refers to patterns of relative genetic similarity among human individuals and populations, as well as the wide range of scientific and statistical methods used to study this aspect of human genetic variation.
Sagiv Shifman is an Israeli scientist, professor in the field of neurogenetics at the Alexander Silberman Institute of Life Sciences, The Hebrew University of Jerusalem. He holds the Arnold and Bess Zeldich Ungerman chair in Neurobiology.
References
- ↑ Zimmer, Carl (2018-08-22). "A Blended Family: Her Mother Was Neanderthal, Her Father Something Else Entirely". The New York Times. ISSN 0362-4331 . Retrieved 2019-04-05.
- ↑ Dvorsky, George (15 March 2018). "Ancient Human Groups Mated With the Mysterious Denisovans at Least Twice". Gizmodo. Retrieved 2019-04-05.
- ↑ "Mysterious Denisovans interbred with modern humans more than once". CNN. 16 March 2018. Retrieved 2018-03-16.
- ↑ "Modern humans interbred with Denisovans twice in history" . Retrieved 2018-03-15.
- ↑ Browning, Sharon (1999). Monte Carlo likelihood calculation for identity by descent data (Thesis thesis).
- ↑ "Browning CV" (PDF). Retrieved December 19, 2021.
- ↑ Browning, Sharon R. (2006-06-01). "Multilocus Association Mapping Using Variable-Length Markov Chains". The American Journal of Human Genetics. 78 (6): 903–913. doi:10.1086/503876. ISSN 0002-9297. PMC 1474089 . PMID 16685642.
- ↑ Browning, Sharon R.; Browning, Brian L. (2007). "Rapid and Accurate Haplotype Phasing and Missing-Data Inference for Whole-Genome Association Studies By Use of Localized Haplotype Clustering". The American Journal of Human Genetics. 81 (5): 1084–1097. doi:10.1086/521987. PMC 2265661 . PMID 17924348.
- ↑ Browning, Sharon R. (2008). "Missing data imputation and haplotype phase inference for genome-wide association studies". Human Genetics. 124 (5): 439–450. doi:10.1007/s00439-008-0568-7. ISSN 0340-6717. PMC 2731769 . PMID 18850115.
- ↑ Madsen, Bo Eskerod; Browning, Sharon R. (2009-02-13). Schork, Nicholas J. (ed.). "A Groupwise Association Test for Rare Mutations Using a Weighted Sum Statistic". PLOS Genetics. 5 (2): e1000384. doi: 10.1371/journal.pgen.1000384 . ISSN 1553-7404. PMC 2633048 . PMID 19214210.
- ↑ Browning, Sharon R.; Browning, Brian L. (2012-12-15). "Identity by Descent Between Distant Relatives: Detection and Applications". Annual Review of Genetics. 46 (1): 617–633. doi:10.1146/annurev-genet-110711-155534. ISSN 0066-4197. PMID 22994355.
- ↑ Browning, Sharon R.; Browning, Brian L. (2010). "High-Resolution Detection of Identity by Descent in Unrelated Individuals". The American Journal of Human Genetics. 86 (4): 526–539. doi:10.1016/j.ajhg.2010.02.021. PMC 2850444 . PMID 20303063.
- ↑ Harris, Daniel N.; Kessler, Michael D.; Shetty, Amol C.; Weeks, Daniel E.; Minster, Ryan L.; Browning, Sharon; Cochrane, Ethan E.; Deka, Ranjan; Hawley, Nicola L.; Reupena, Muagututi‘a Sefuiva; Naseri, Take (2020-04-28). "Evolutionary history of modern Samoans". Proceedings of the National Academy of Sciences. 117 (17): 9458–9465. Bibcode:2020PNAS..117.9458H. doi: 10.1073/pnas.1913157117 . ISSN 0027-8424. PMC 7196816 . PMID 32291332.
- ↑ "Researchers use 21st century genomics to estimate Samoan population dynamics over 3,000 years". Brown University. Retrieved 2021-12-23.
- ↑ "Multiple lines of mysterious ancient humans interbred with us". Science. 2019-04-11. Retrieved 2021-12-23.
- ↑ Guarino, Ben (March 15, 2018). "Humans bred with this mysterious species more than once, new study shows". Washington Post. ISSN 0190-8286 . Retrieved 2021-12-23.
- ↑ Browning, Sharon R.; Browning, Brian L.; Zhou, Ying; Tucci, Serena; Akey, Joshua M. (2018). "Analysis of Human Sequence Data Reveals Two Pulses of Archaic Denisovan Admixture". Cell. 173 (1): 53–61.e9. doi:10.1016/j.cell.2018.02.031. PMC 5866234 . PMID 29551270.