
Staphylococcus aureus is a Gram-positive, round-shaped bacterium that is a member of the Firmicutes, and it is a usual member of the microbiota of the body, frequently found in the upper respiratory tract and on the skin. It is often positive for catalase and nitrate reduction and is a facultative anaerobe that can grow without the need for oxygen. Although S. aureus usually acts as a commensal of the human microbiota it can also become an opportunistic pathogen, being a common cause of skin infections including abscesses, respiratory infections such as sinusitis, and food poisoning. Pathogenic strains often promote infections by producing virulence factors such as potent protein toxins, and the expression of a cell-surface protein that binds and inactivates antibodies. The emergence of antibiotic-resistant strains of S. aureus such as methicillin-resistant S. aureus (MRSA) is a worldwide problem in clinical medicine. Despite much research and development, no vaccine for S. aureus has been approved.
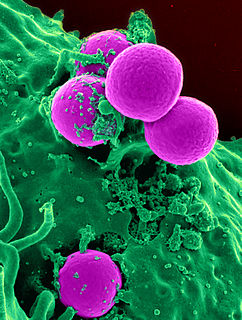
Methicillin-resistant Staphylococcus aureus (MRSA) refers to a group of Gram-positive bacteria that are genetically distinct from other strains of Staphylococcus aureus. MRSA is responsible for several difficult-to-treat infections in humans. MRSA is any strain of S. aureus that has developed or acquired a multiple drug resistance to beta-lactam antibiotics. Beta-lactam (β-lactam) antibiotics are a broad-spectrum group that include some penams and cephems such as the cephalosporins. Strains unable to resist these antibiotics are classified as methicillin-susceptible S. aureus, or MSSA.

Vancomycin-resistant Staphylococcus aureus (VRSA) are strains of Staphylococcus aureus that have become resistant to the glycopeptide antibiotic vancomycin.

Staphylococcus haemolyticus is a member of the coagulase-negative staphylococci (CoNS). It is part of the skin flora of humans, and its largest populations are usually found at the axillae, perineum, and inguinal areas. S. haemolyticus also colonizes primates and domestic animals. It is a well-known opportunistic pathogen, and is the second-most frequently isolated CoNS. Infections can be localized or systemic, and are often associated with the insertion of medical devices. The highly antibiotic-resistant phenotype and ability to form biofilms make S. haemolyticus a difficult pathogen to treat. Its most closely-related species if Staphylococcus borealis.
RNAIII is a stable 514 nt regulatory RNA transcribed by the P3 promoter of the Staphylococcus aureus quorum-sensing agr system ). It is the major effector of the agr regulon, which controls the expression of many S. aureus genes encoding exoproteins and cell wall associated proteins plus others encoding regulatory proteins The RNAIII transcript also encodes the 26 amino acid δ-haemolysin peptide (Hld). RNAIII contains many stem loops, most of which match the Shine-Dalgarno sequence involved in translation initiation of the regulated genes. Some of these interactions are inhibitory, others stimulatory; among the former is the regulatory protein Rot. In vitro, RNAIII is expressed post exponentially, inhibiting translation of the surface proteins, notably protein A, while stimulating that of the exoproteins, many of which are tissue-degrading enzymes or cytolysins. Among the latter is the important virulence factor, α-hemolysin (Hla), whose translation RNAIII activates by preventing the formation of an inhibitory foldback loop in the hla mRNA leader.
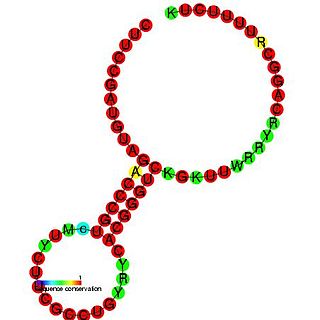
RydC is a bacterial non-coding RNA. RydC is thought to regulate a mRNA, yejABEF, which encodes an ABC transporter protein. RydC is known to bind the Hfq protein, which causes a conformational change in the RNA molecule. The Hfq/RydC complex is then thought to bind to the target mRNA and induce its degradation.

EF-G is a prokaryotic elongation factor involved in protein translation. As a GTPase, EF-G catalyzes the movement (translocation) of transfer RNA (tRNA) and messenger RNA (mRNA) through the ribosome.

Listeria monocytogenes is a gram positive bacterium and causes many food-borne infections such as Listeriosis. This bacteria is ubiquitous in the environment where it can act as either a saprophyte when free living within the environment or as a pathogen when entering a host organism. Many non-coding RNAs have been identified within the bacteria genome where several of these have been classified as novel non-coding RNAs and may contribute to pathogenesis.
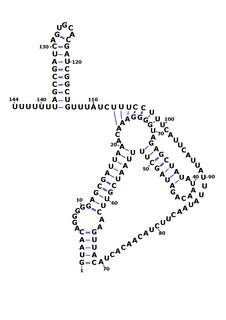
RsaOG is a non-coding RNA that was discovered in the pathogenic bacteria Staphylococcus aureus N315 using a large scale computational screening based on phylogenetic profiling. It was first identified, but not named, in 2005. RsaOG has since been identified in other strains of Staphylococcus aureus under the name of RsaI, it has also been discovered in other members of the Staphylococcus genus but in no other bacteria.

Rhein, also known as cassic acid, is a substance in the anthraquinone group obtained from rhubarb. Like all such substances, rhein is a cathartic. Rhein is commonly found as a glycoside such as rhein-8-glucoside or glucorhein. Rhein was first isolated in 1895. It is found in rhubarb species like Rheum undulatum and Rheum palmatum as well as in Cassia reticulata.
Bacterial small RNAs (sRNA) are small RNAs produced by bacteria; they are 50- to 500-nucleotide non-coding RNA molecules, highly structured and containing several stem-loops. Numerous sRNAs have been identified using both computational analysis and laboratory-based techniques such as Northern blotting, microarrays and RNA-Seq in a number of bacterial species including Escherichia coli, the model pathogen Salmonella, the nitrogen-fixing alphaproteobacterium Sinorhizobium meliloti, marine cyanobacteria, Francisella tularensis, Streptococcus pyogenes, the pathogen Staphylococcus aureus, and the plant pathogen Xanthomonas oryzae pathovar oryzae. Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNA or protein, and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress response, and structure.
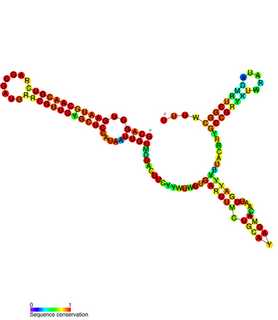
FnrS RNA is a family of Hfq-binding small RNA whose expression is upregulated in response to anaerobic conditions. It is named FnrS because its expression is strongly dependent on fumarate and nitrate reductase regulator (FNR), a direct oxygen availability sensor.
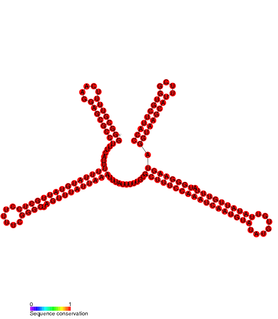
In molecular biology SprD is a non-coding RNA expressed on pathogenicity islands in Staphylococcus aureus. It was identified in silico along with a number of other sRNAs (SprA-G) through microarray analysis which were confirmed using a Northern blot. SprD has been found to significantly contribute to causing disease in an animal model.

A toxin-antitoxin system is a set of two or more closely linked genes that together encode both a "toxin" protein and a corresponding "antitoxin". Toxin-antitoxin systems are widely distributed in prokaryotes, and organisms often have them in multiple copies. When these systems are contained on plasmids – transferable genetic elements – they ensure that only the daughter cells that inherit the plasmid survive after cell division. If the plasmid is absent in a daughter cell, the unstable antitoxin is degraded and the stable toxic protein kills the new cell; this is known as 'post-segregational killing' (PSK).
Rsa RNAs are non-coding RNAs found in the bacterium Staphylococcus aureus. The shared name comes from their discovery, and does not imply homology. Bioinformatics scans identified the 16 Rsa RNA families named RsaA-K and RsaOA-OG. Others, RsaOH-OX, were found thanks to an RNomic approach. Although the RNAs showed varying expression patterns, many of the newly discovered RNAs were shown to be Hfq-independent and most carried a C-rich motif (UCCC).
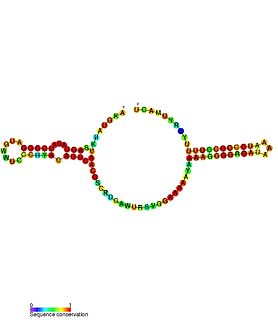
Escherichia coli contains a number of small RNAs located in intergenic regions of its genome. The presence of at least 55 of these has been verified experimentally. 275 potential sRNA-encoding loci were identified computationally using the QRNA program. These loci will include false positives, so the number of sRNA genes in E. coli is likely to be less than 275. A computational screen based on promoter sequences recognised by the sigma factor sigma 70 and on Rho-independent terminators predicted 24 putative sRNA genes, 14 of these were verified experimentally by northern blotting. The experimentally verified sRNAs included the well characterised sRNAs RprA and RyhB. Many of the sRNAs identified in this screen, including RprA, RyhB, SraB and SraL, are only expressed in the stationary phase of bacterial cell growth. A screen for sRNA genes based on homology to Salmonella and Klebsiella identified 59 candidate sRNA genes. From this set of candidate genes, microarray analysis and northern blotting confirmed the existence of 17 previously undescribed sRNAs, many of which bind to the chaperone protein Hfq and regulate the translation of RpoS. UptR sRNA transcribed from the uptR gene is implicated in suppressing extracytoplasmic toxicity by reducing the amount of membrane-bound toxic hybrid protein.
16S rRNA (guanine1405-N7)-methyltransferase (EC 2.1.1.179, methyltransferase Sgm, m7G1405 Mtase, Sgm Mtase, Sgm, sisomicin-gentamicin methyltransferase, sisomicin-gentamicin methylase, GrmA, RmtB, RmtC, ArmA) is an enzyme with systematic name S-adenosyl-L-methionine:16S rRNA (guanine1405-N7)-methyltransferase. This enzyme catalyses the following chemical reaction
Staphylococcus pseudintermedius is a gram positive coccus bacteria of the genus Staphylococcus found worldwide. It is primarily a pathogen for domestic animals, but has been known to affect humans as well.S. pseudintermedius is an opportunistic pathogen that secretes immune modulating virulence factors, has many adhesion factors, and the potential to create biofilms, all of which help to determine the pathogenicity of the bacterium. Diagnoses of Staphylococcus pseudintermedius have traditionally been made using cytology, plating, and biochemical tests. More recently, molecular technologies like MALDI-TOF, DNA hybridization and PCR have become preferred over biochemical tests for their more rapid and accurate identifications. This includes the identification and diagnosis of antibiotic resistant strains.
The SprA1/SPrA1as toxin/antitoxin system identified in Staphylococcus aureus, belongs to the Type I system encoding toxin protein: SprA1 and antitoxin RNA: SprA1as. The SprA1as postranscriptionally regulates SprA1 encoding small membrane damaging protein PepA1.













